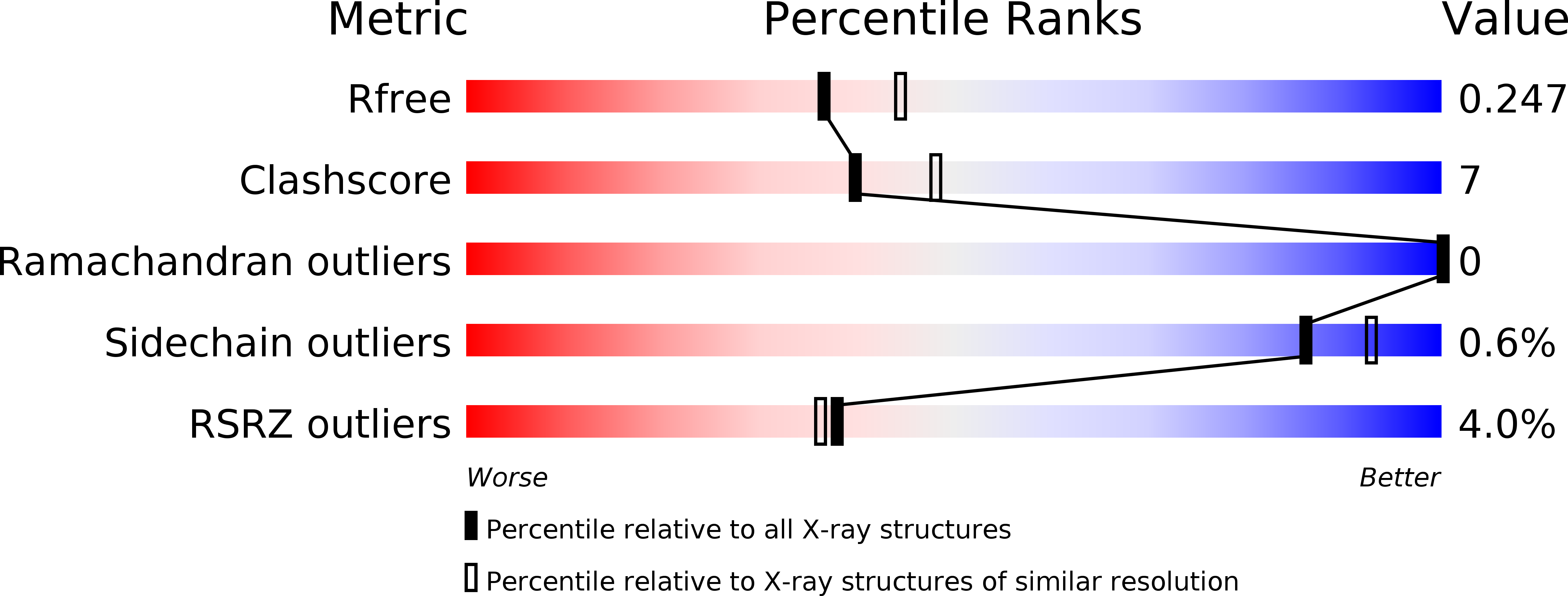
The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Klunemann, T., Henke, S., Blankenfeldt, W.(2020) Acta Crystallogr D Struct Biol 76: 375-384
- PubMed: 32254062
- DOI: https://doi.org/10.1107/S2059798320003101
- Primary Citation of Related Structures:
6TP9 - PubMed Abstract:
Monoheme c-type cytochromes are important electron transporters in all domains of life. They possess a common fold hallmarked by three α-helices that surround a covalently attached heme. An intriguing feature of many monoheme c-type cytochromes is their capacity to form oligomers by exchanging at least one of their α-helices, which is often referred to as 3D domain swapping. Here, the crystal structure of NirC, a c-type cytochrome co-encoded with other proteins involved in nitrite reduction by the opportunistic pathogen Pseudomonas aeruginosa, has been determined. The crystals diffracted anisotropically to a maximum resolution of 2.12 Å (spherical resolution of 2.83 Å) and initial phases were obtained by Fe-SAD phasing, revealing the presence of 11 NirC chains in the asymmetric unit. Surprisingly, these protomers arrange into one monomer and two different types of 3D domain-swapped dimers, one of which shows pronounced asymmetry. While the simultaneous observation of monomers and dimers probably reflects the interplay between the high protein concentration required for crystallization and the structural plasticity of monoheme c-type cytochromes, the identification of conserved structural motifs in the monomer together with a comparison with similar proteins may offer new leads to unravel the unknown function of NirC.
Organizational Affiliation:
Structure and Function of Proteins, Helmholtz Centre for Infection Research, Inhoffenstrasse 7, 38124 Braunschweig, Germany.