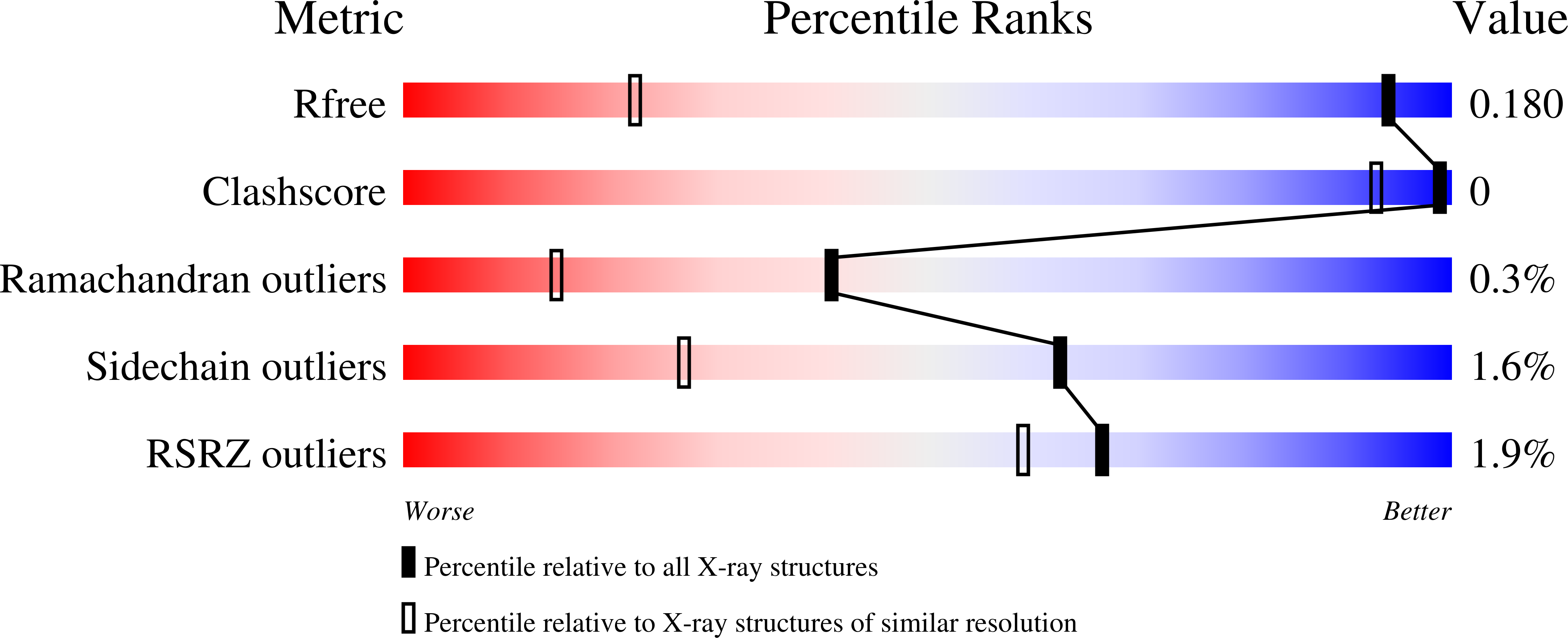
Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Miarzlou, D.A., Leisinger, F., Joss, D., Haussinger, D., Seebeck, F.P.(2019) Chem Sci 10: 7049-7058
- PubMed: 31588272
- DOI: https://doi.org/10.1039/c9sc01723b
- Primary Citation of Related Structures:
6S07 - PubMed Abstract:
The formylglycine generating enzyme (FGE) catalyzes oxidative conversion of specific peptidyl-cysteine residues to formylglycine. FGE mediates O 2 -activation and hydrogen-atom abstraction in an active site that contains Cu(i) coordinated to two cysteine residues. Similar coordination geometries are common among copper-sensing transcription factors and copper-chaperone but are unprecedented among copper-dependent oxidases. To examine the mechanism of this unusual catalyst we determined the 1.04 Å structure of FGE from Thermomonospora curvata in complex with copper and a cysteine-containing peptide substrate. This structure unveils a network of four crystallographic waters and two active site residues that form a highly acidic O 2 -binding pocket juxtaposed to the trigonal planar tris-cysteine coordinated Cu(i) center. Comparison with structures of FGE in complex with Ag(i) and Cd(ii) combined with evidence from NMR spectroscopy and kinetic observations highlight several structural changes that are induced by substrate binding and prime the enzyme for O 2 -binding and subsequent activation.
Organizational Affiliation:
Department of Chemistry , University of Basel , Mattenstrasse 24a , Basel 4002 , Switzerland . Email: florian.seebeck@unibas.ch ; Tel: +41 612071143.