The structural basis for an on-off switch controlling G beta gamma-mediated inhibition of TRPM3 channels.
Behrendt, M., Gruss, F., Enzeroth, R., Dembla, S., Zhao, S., Crassous, P.A., Mohr, F., Nys, M., Louros, N., Gallardo, R., Zorzini, V., Wagner, D., Economou, A., Rousseau, F., Schymkowitz, J., Philipp, S.E., Rohacs, T., Ulens, C., Oberwinkler, J.(2020) Proc Natl Acad Sci U S A 117: 29090-29100
- PubMed: 33122432
- DOI: https://doi.org/10.1073/pnas.2001177117
- Primary Citation of Related Structures:
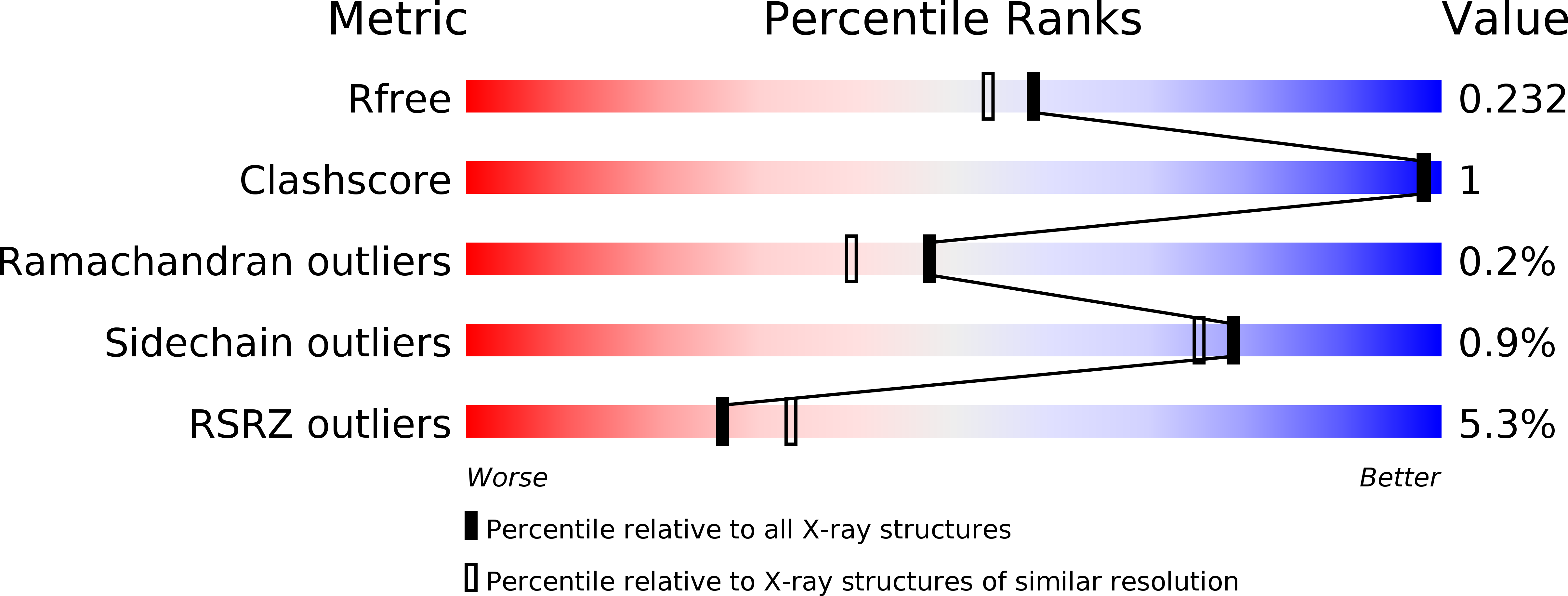
6RMV - PubMed Abstract:
TRPM3 channels play important roles in the detection of noxious heat and in inflammatory thermal hyperalgesia. The activity of these ion channels in somatosensory neurons is tightly regulated by µ-opioid receptors through the signaling of Gβγ proteins, thereby reducing TRPM3-mediated pain. We show here that Gβγ directly binds to a domain of 10 amino acids in TRPM3 and solve a cocrystal structure of this domain together with Gβγ. Using these data and mutational analysis of full-length proteins, we pinpoint three amino acids in TRPM3 and their interacting partners in Gβ 1 that are individually necessary for TRPM3 inhibition by Gβγ. The 10-amino-acid Gβγ-interacting domain in TRPM3 is subject to alternative splicing. Its inclusion in or exclusion from TRPM3 channel proteins therefore provides a mechanism for switching on or off the inhibitory action that Gβγ proteins exert on TRPM3 channels.
Organizational Affiliation:
Institut für Physiologie und Pathophysiologie, Philipps-Universität Marburg, 35037 Marburg, Germany.