Bioinspired Hybrid Fluorescent Ligands for the FK1 Domain of FKBP52.
de la Sierra-Gallay, I.L., Belnou, M., Chambraud, B., Genet, M., van Tilbeurgh, H., Aumont-Nicaise, M., Desmadril, M., Baulieu, E.E., Jacquot, Y., Byrne, C.(2020) J Med Chem 63: 10330-10338
- PubMed: 32866001
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00825
- Primary Citation of Related Structures:
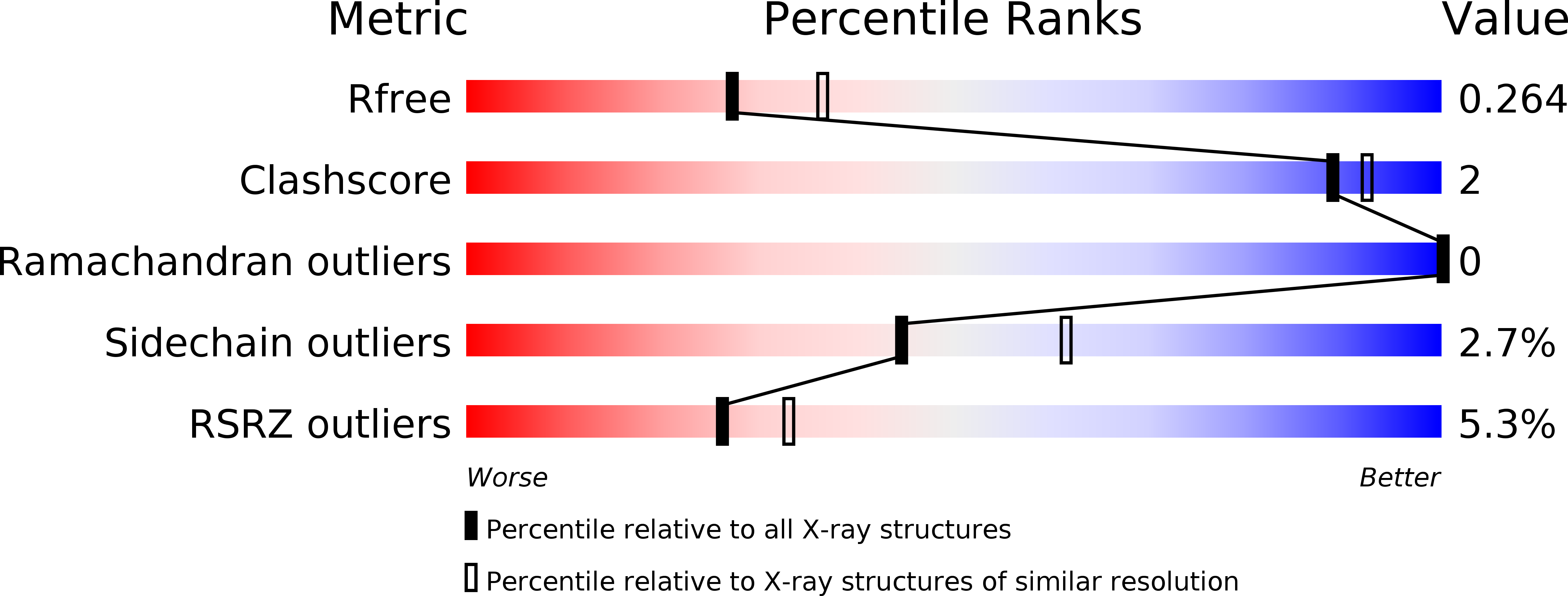
6RCY - PubMed Abstract:
The protein FKBP52 is a steroid hormone receptor coactivator likely involved in neurodegenerative disease. A series of small, water-soluble, bioinspired, pseudopeptidic fluorescent ligands for the FK1 domain of this protein are described. The design is such that engulfing of the ligand in the pocket of this domain is accompanied by hydrogen-bonding of the dansyl chromophore which functions as both an integral part of the ligand and a fluorescent reporter. Binding is concomitant with a significant wavelength shift and an enhancement of the ligand fluorescence signal. Excitation of FK1 domain native tryptophan residues in the presence of bound ligand results in Förster resonance energy transfer. Variation of key ligand residues within the short sequence was undertaken, and the interaction of the resulting library with the protein was measured by techniques including isothermal calorimetry analysis, fluorescence, and FRET quenching, and a range of K d values were determined. Cocrystallization of a protein ligand complex at 2.30 Å resolution provided detailed information at the atomic scale, while also providing insight into native substrate binding.
Organizational Affiliation:
Institut de Biologie Intégrative de la Cellule (I2BC), CNRS UMR9198, Université Paris-Saclay, Université Paris-Sud, 91405 Orsay, France.