How to Computationally Stack the Deck for Hit-to-Lead Generation: In Silico Molecular Interaction Energy Profiling for de Novo siRNA Guide Strand Surrogate Selection.
Greenidge, P.A., Blommers, M.J.J., Priestle, J.P., Hunziker, J.(2019) J Chem Inf Model 59: 1897-1908
- PubMed: 31021613
- DOI: https://doi.org/10.1021/acs.jcim.8b00892
- Primary Citation of Related Structures:
6RA4 - PubMed Abstract:
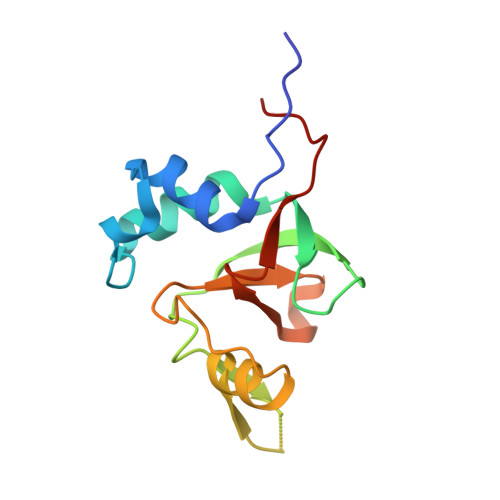
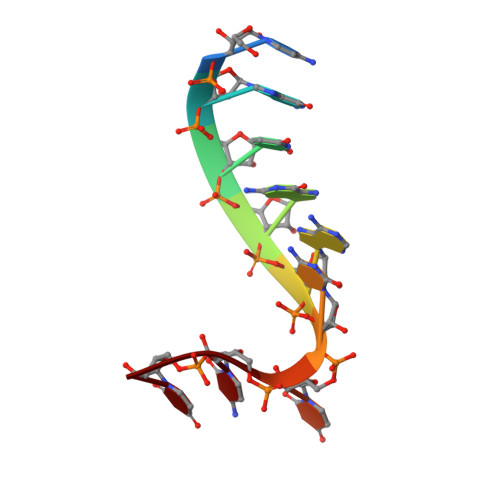
The Argonaute-2 protein is part of the RNA-induced silencing complex (RISC) and anchors the guide strand of the small interfering RNA (siRNA). The 3'-end of the RNA contains two unpaired nucleotides (3'-overhang) that interact with the PAZ (PIWI/Argonaute/Zwille) domain of the protein. Theoretical and experimental evidence points toward a direct connection between the PAZ/3'-overhang binding affinity and siRNA's potency and specificity. Among the challenges to overcome when deploying siRNA molecules as therapeutics are their ready degradation under physiological conditions and off-target effects. It has been demonstrated that nuclease resistance can be improved via replacement of the dinucleotide overhang by small molecules which retain the interactions of the RNA guide strand with the PAZ domain. Most commonly, nucleotide analogues are used to substitute the siRNA overhang. However, in this study we adopt a de novo approach to its modification. The X-ray structure of human Argonaute-2 PAZ domain served to perform virtual screening and molecular interaction energy profiling (i.e., decomposition of the force field calculated protein-ligand interaction energies) of tailored-to-purpose fragment libraries. The binding of fragments to the PAZ domain was validated experimentally by NMR spectroscopy. The in silico guided protocol led to the efficient discovery of a number of PAZ domain ligands with affinities comparable to that of a reference dinucleotide (UpU, K d = 33 μM). Originally starting from a generic fragment library, hits progress from 930 μM down to 14 μM within three iterations for the fragments selected via in silico molecular interaction energy profiling from a bespoke library. These dinucleotide siRNA guide strand surrogates represent potential new siRNA-based therapeutics (when attached to siRNA to form bioconjugates) featuring improved efficacy, specificity, stability, and cellular uptake. This project yielded a portfolio of seven patent applications, four of which have been granted to date.
Organizational Affiliation:
Novartis Institutes for BioMedical Research (NIBR), Novartis Pharma AG , Forum 1, Novartis Campus, Fabrikstrasse 2 , CH-4056 Basel , Switzerland.