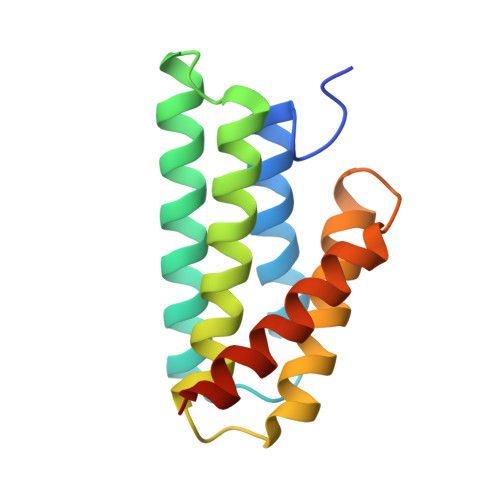
Structure of the N-terminal domain of Euprosthenops australis dragline silk suggests that conversion of spidroin dope to spider silk involves a conserved asymmetric dimer intermediate.
Jiang, W., Askarieh, G., Shkumatov, A., Hedhammar, M., Knight, S.D.(2019) Acta Crystallogr D Struct Biol 75: 618-627
- PubMed: 31282471
- DOI: https://doi.org/10.1107/S2059798319007253
- Primary Citation of Related Structures:
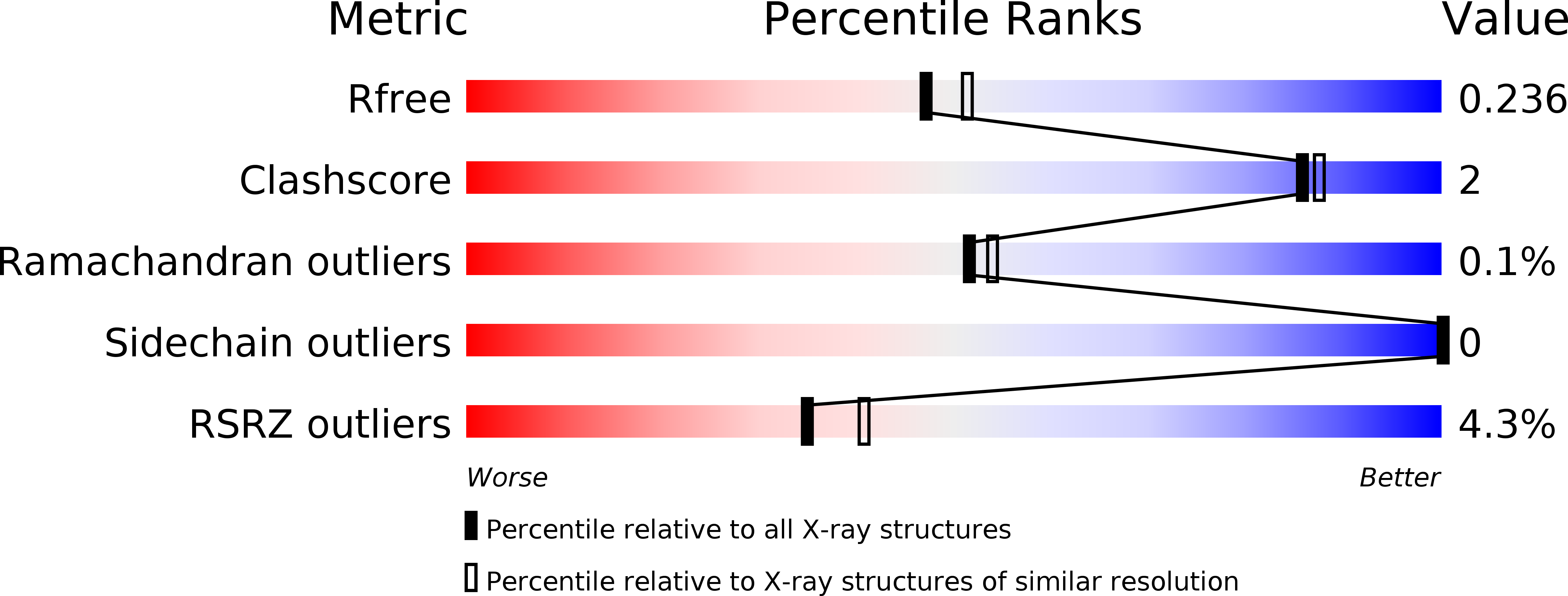
6R9D - PubMed Abstract:
Spider silk is a biomaterial with exceptional mechanical toughness, and there is great interest in developing biomimetic methods to produce engineered spider silk-based materials. However, the mechanisms that regulate the conversion of spider silk proteins (spidroins) from highly soluble dope into silk are not completely understood. The N-terminal domain (NT) of Euprosthenops australis dragline silk protein undergoes conformational and quaternary-structure changes from a monomer at a pH above 7 to a homodimer at lower pH values. Conversion from the monomer to the dimer requires the protonation of three conserved glutamic acid residues, resulting in a low-pH `locked' dimer stabilized by symmetric electrostatic interactions at the poles of the dimer. The detailed molecular events during this transition are still unresolved. Here, a 2.1 Å resolution crystal structure of an NT T61A mutant in an alternative, asymmetric, dimer form in which the electrostatic interactions at one of the poles are dramatically different from those in symmetrical dimers is presented. A similar asymmetric dimer structure from dragline silk of Nephila clavipes has previously been described. It is suggested that asymmetric dimers represent a conserved intermediate state in spider silk formation, and a revised `lock-and-trigger' mechanism for spider silk formation is presented.
Organizational Affiliation:
Department of Cell and Molecular Biology, Uppsala University, Biomedical Center, PO Box 596, SE-751 24 Uppsala, Sweden.