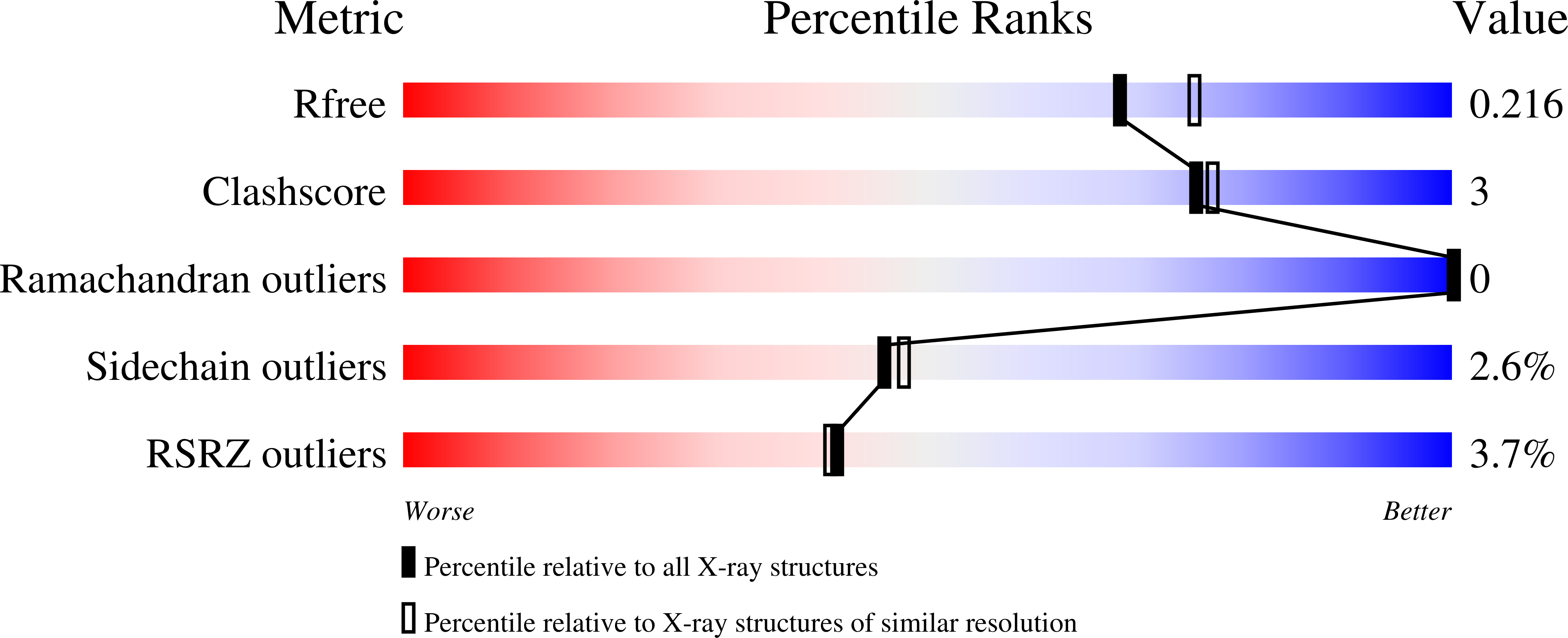
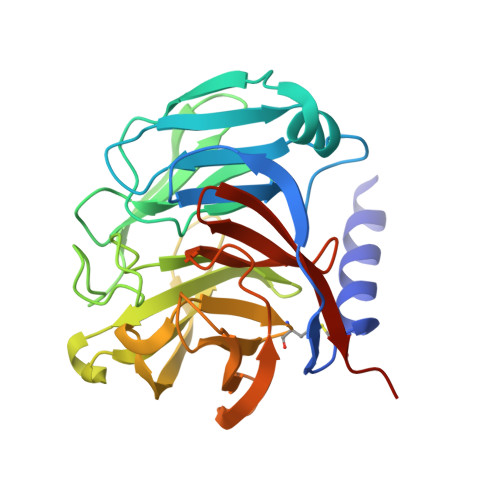
Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
Pronker, M.F., van den Hoek, H., Janssen, B.J.C.(2019) BMC Mol Cell Biol 20: 50-50
- PubMed: 31726976
- DOI: https://doi.org/10.1186/s12860-019-0232-1
- Primary Citation of Related Structures:
6QHJ, 6QM3 - PubMed Abstract:
Olfactomedin-1 (Olfm1; also known as Noelin or Pancortin) is a highly-expressed secreted brain and retina protein and its four isoforms have different roles in nervous system development and function. Structural studies showed that the long Olfm1 isoform BMZ forms a disulfide-linked tetramer with a V-shaped architecture. The tips of the Olfm1 "V" each consist of two C-terminal β-propeller domains that enclose a calcium binding site. Functional characterisation of Olfm1 may be aided by new biochemical tools derived from these core structural elements. Here we present the production, purification and structural analysis of three novel monomeric, dimeric and tetrameric forms of mammalian Olfm1 for functional studies. We characterise these constructs structurally by high-resolution X-ray crystallography and small-angle X-ray scattering. The crystal structure of the Olfm1 β-propeller domain (to 1.25 Å) represents the highest-resolution structure of an olfactomedin family member to date, revealing features such as a hydrophilic tunnel containing water molecules running into the core of the domain where the calcium binding site resides. The shorter Olfactomedin-1 isoform BMY is a disulfide-linked tetramer with a shape similar to the corresponding region in the longer BMZ isoform. These recombinantly-expressed protein tools should assist future studies, for example of biophysical, electrophysiological or morphological nature, to help elucidate the functions of Olfm1 in the mature mammalian brain. The control over the oligomeric state of Olfm1 provides a firm basis to better understand the role of Olfm1 in the (trans-synaptic) tethering or avidity-mediated clustering of synaptic receptors such as post-synaptic AMPA receptors and pre-synaptic amyloid precursor protein. In addition, the variation in domain composition of these protein tools provides a means to dissect the Olfm1 regions important for receptor binding.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Division of Neurobiology, Francis Crick Avenue, Cambridge, CB2 0QH, UK. m.f.pronker@uu.nl.