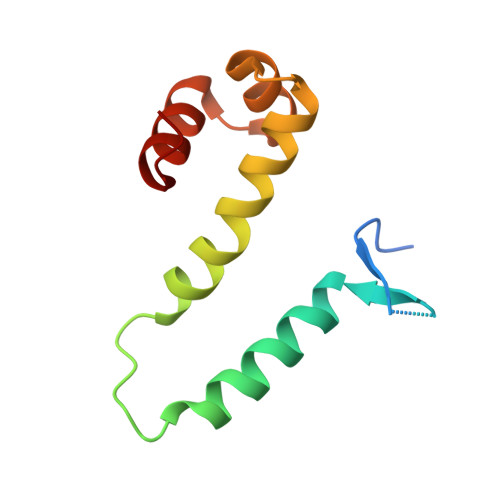
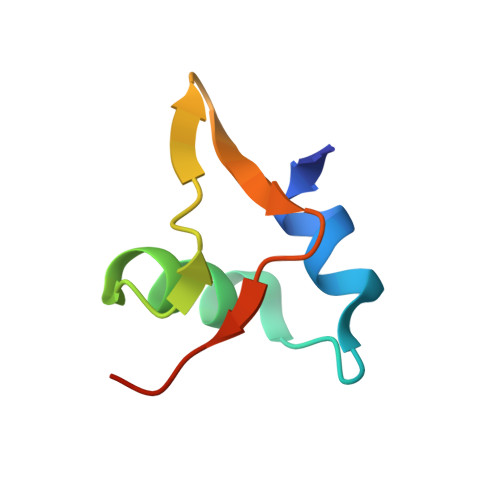
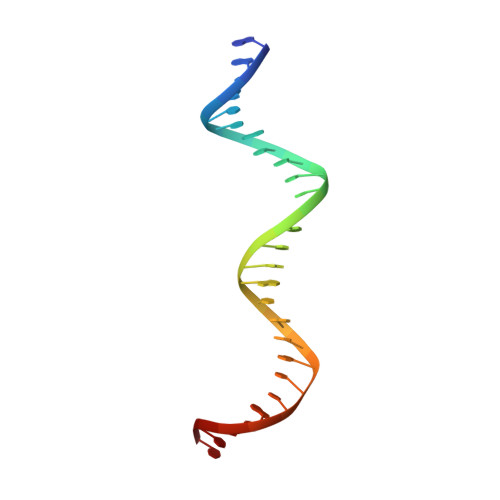
Cooperative DNA binding by proteins through DNA shape complementarity.
Hancock, S.P., Cascio, D., Johnson, R.C.(2019) Nucleic Acids Res 47: 8874-8887
- PubMed: 31616952
- DOI: https://doi.org/10.1093/nar/gkz642
- Primary Citation of Related Structures:
6P0S, 6P0T, 6P0U - PubMed Abstract:
Localized arrays of proteins cooperatively assemble onto chromosomes to control DNA activity in many contexts. Binding cooperativity is often mediated by specific protein-protein interactions, but cooperativity through DNA structure is becoming increasingly recognized as an additional mechanism. During the site-specific DNA recombination reaction that excises phage λ from the chromosome, the bacterial DNA architectural protein Fis recruits multiple λ-encoded Xis proteins to the attR recombination site. Here, we report X-ray crystal structures of DNA complexes containing Fis + Xis, which show little, if any, contacts between the two proteins. Comparisons with structures of DNA complexes containing only Fis or Xis, together with mutant protein and DNA binding studies, support a mechanism for cooperative protein binding solely by DNA allostery. Fis binding both molds the minor groove to potentiate insertion of the Xis β-hairpin wing motif and bends the DNA to facilitate Xis-DNA contacts within the major groove. The Fis-structured minor groove shape that is optimized for Xis binding requires a precisely positioned pyrimidine-purine base-pair step, whose location has been shown to modulate minor groove widths in Fis-bound complexes to different DNA targets.
Organizational Affiliation:
Department of Biological Chemistry, David Geffen School of Medicine at the University of California at Los Angeles, Los Angeles, CA 90095-1737, USA.