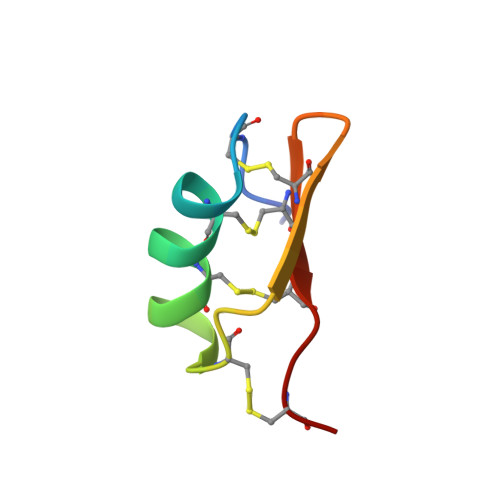
Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Luna-Ramirez, K., Csoti, A., McArthur, J.R., Chin, Y.K.Y., Anangi, R., Najera, R.D.C., Possani, L.D., King, G.F., Panyi, G., Yu, H., Adams, D.J., Finol-Urdaneta, R.K.(2020) Biochem Pharmacol 174: 113782-113782
- PubMed: 31881193
- DOI: https://doi.org/10.1016/j.bcp.2019.113782
- Primary Citation of Related Structures:
6MZT - PubMed Abstract:
Urotoxin (α-KTx 6), a peptide from venom of the Australian scorpion Urodacus yaschenkoi, is the most potent inhibitor of Kv1.2 described to date (IC 50 = 160 pM). The native peptide also inhibits Kv1.1, Kv1.3 and KCa3.1 with nanomolar affinity but its low abundance in venom precluded further studies of its actions. Here we produced recombinant Urotoxin (rUro) and characterized the molecular determinants of Kv1 channel inhibition. The 3D structure of rUro determined using NMR spectroscopy revealed a canonical cysteine-stabilised α/β (CSα/β) fold. Functional assessment of rUro using patch-clamp electrophysiology revealed the importance of C-terminal amidation for potency against Kv1.1-1.3 and Kv1.5. Neutralization of the putative pore-blocking K25 residue in rUro by mutation to Ala resulted in a major decrease in rUro potency against all Kv channels tested, without perturbing the toxin's structure. Reciprocal mutations in the pore of Uro-sensitive Kv1.2 and Uro-resistant Kv1.5 channels revealed a direct interaction between Urotoxin and the Kv channel pore. Our experimental work supports postulating a mechanism of action in which occlusion of the permeation pathway by the K25 residue in Urotoxin is the basis of its Kv1 inhibitory activity. Docking analysis was consistent with occlusion of the pore by K25 and the requirement of a small, non-charged amino acid in the Kv1 channel vestibule to facilitate toxin-channel interactions. Finally, computational studies revealed key interactions between the amidated C-terminus of Urotoxin and a conserved Asp residue in the turret of Kv1 channels, offering a potential rationale for potency differences between native and recombinant Urotoxin.
Organizational Affiliation:
Illawarra Health and Medical Research Institute (IHMRI), University of Wollongong, Wollongong, NSW 2522, Australia. Electronic address: ramirezk@uow.edu.au.