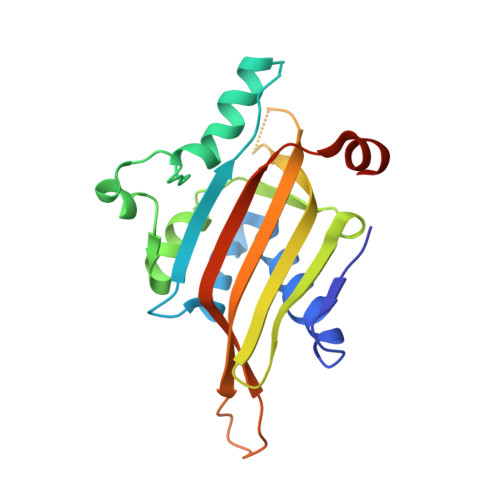
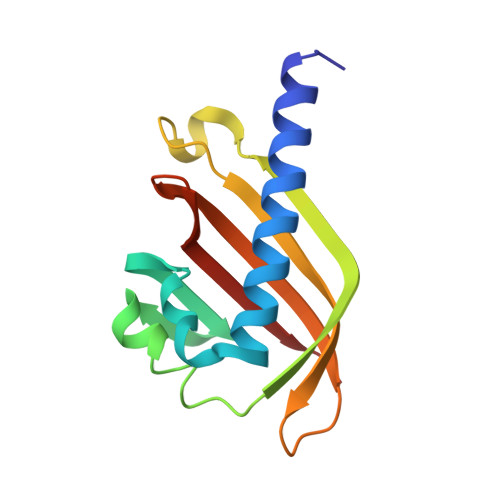
The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation.
Batki, J., Schnabl, J., Wang, J., Handler, D., Andreev, V.I., Stieger, C.E., Novatchkova, M., Lampersberger, L., Kauneckaite, K., Xie, W., Mechtler, K., Patel, D.J., Brennecke, J.(2019) Nat Struct Mol Biol 26: 720-731
- PubMed: 31384064
- DOI: https://doi.org/10.1038/s41594-019-0270-6
- Primary Citation of Related Structures:
6MRK, 6OPF - PubMed Abstract:
The PIWI-interacting RNA (piRNA) pathway protects genome integrity in part through establishing repressive heterochromatin at transposon loci. Silencing requires piRNA-guided targeting of nuclear PIWI proteins to nascent transposon transcripts, yet the subsequent molecular events are not understood. Here, we identify SFiNX (silencing factor interacting nuclear export variant), an interdependent protein complex required for Piwi-mediated cotranscriptional silencing in Drosophila. SFiNX consists of Nxf2-Nxt1, a gonad-specific variant of the heterodimeric messenger RNA export receptor Nxf1-Nxt1 and the Piwi-associated protein Panoramix. SFiNX mutant flies are sterile and exhibit transposon derepression because piRNA-loaded Piwi is unable to establish heterochromatin. Within SFiNX, Panoramix recruits heterochromatin effectors, while the RNA binding protein Nxf2 licenses cotranscriptional silencing. Our data reveal how Nxf2 might have evolved from an RNA transport receptor into a cotranscriptional silencing factor. Thus, NXF variants, which are abundant in metazoans, can have diverse molecular functions and might have been coopted for host genome defense more broadly.
Organizational Affiliation:
Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter, Vienna, Austria.