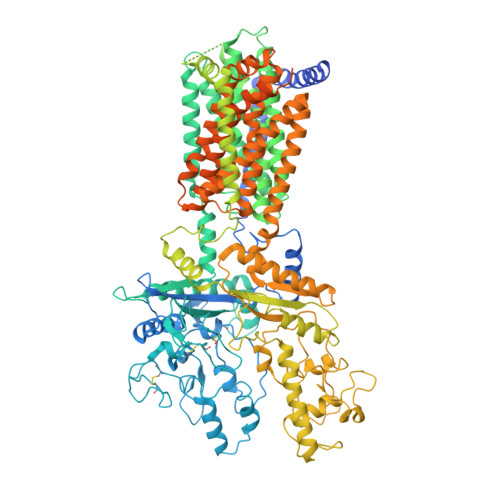
Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Zhang, Y., Bulkley, D.P., Xin, Y., Roberts, K.J., Asarnow, D.E., Sharma, A., Myers, B.R., Cho, W., Cheng, Y., Beachy, P.A.(2018) Cell 175: 1352-1364.e14
- PubMed: 30415841
- DOI: https://doi.org/10.1016/j.cell.2018.10.026
- Primary Citation of Related Structures:
6MG8 - PubMed Abstract:
Hedgehog protein signals mediate tissue patterning and maintenance by binding to and inactivating their common receptor Patched, a 12-transmembrane protein that otherwise would suppress the activity of the 7-transmembrane protein Smoothened. Loss of Patched function, the most common cause of basal cell carcinoma, permits unregulated activation of Smoothened and of the Hedgehog pathway. A cryo-EM structure of the Patched protein reveals striking transmembrane domain similarities to prokaryotic RND transporters. A central hydrophobic conduit with cholesterol-like contents courses through the extracellular domain and resembles that used by other RND proteins to transport substrates, suggesting Patched activity in cholesterol transport. Cholesterol activity in the inner leaflet of the plasma membrane is reduced by PTCH1 expression but rapidly restored by Hedgehog stimulation, suggesting that PTCH1 regulates Smoothened by controlling cholesterol availability.
Organizational Affiliation:
Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA; Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA 94158, USA.