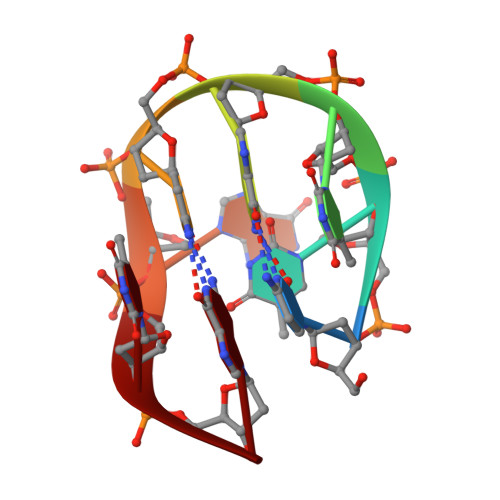
High-Resolution Structures of DNA Minidumbbells Comprising Type II Tetraloops with a Purine Minor Groove Residue.
Ngai, C.K., Lam, S.L., Lee, H.K., Guo, P.(2020) J Phys Chem B 124: 5131-5138
- PubMed: 32484672
- DOI: https://doi.org/10.1021/acs.jpcb.0c03163
- Primary Citation of Related Structures:
6M0B, 6M0C - PubMed Abstract:
Minidumbbell (MDB) is a newly discovered DNA structure formed by native sequences, which serves as a possible structural intermediate causing repeat expansion mutations in the genome and also a functional structural motif in constructing DNA-based molecular switches. Until now, all the reported MDBs containing two adjacent type II tetraloops were formed by pyrimidine-rich sequences 5'-YYYR YYYR-3' (Y and R represent pyrimidine and purine, respectively), wherein the second and sixth residues folded into the minor groove and interacted with each other. In this study, we have conducted a high-resolution nuclear magnetic resonance (NMR) spectroscopic investigation on alternative MDB-forming sequences and discovered that an MDB could also be formed stably with a purine in the minor groove, which has never been observed in any previously reported DNA type II tetraloops. Our refined NMR solution structures of the two MDBs formed by 5'-CTTG C A TG-3' and 5'-CTTG C G TG-3' reveal that the sixth purine residue was driven into the minor groove via base-base stacking with the second thymine residue and adenine stacked better than guanine. The results of our present research work expand the sequence criteria for the formation of MDBs and shed light to explore the significance of MDBs.
Organizational Affiliation:
Department of Chemistry, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong SAR, China.