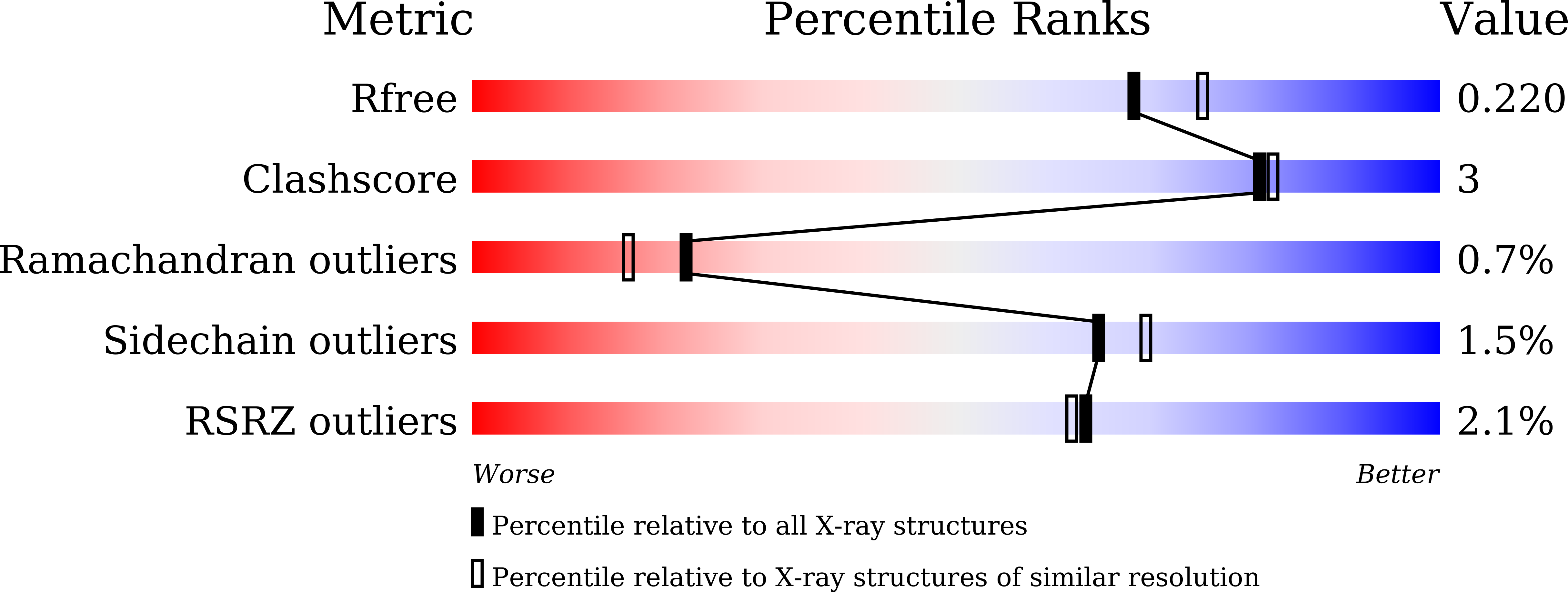
Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Si, Y., Yao, Y., Jaramillo Ayala, G., Li, X., Han, Q., Zhang, W., Xu, X., Tai, G., Mayo, K.H., Zhou, Y., Su, J.(2020) Biochim Biophys Acta Gen Subj 1865: 129755-129755
- PubMed: 33011338
- DOI: https://doi.org/10.1016/j.bbagen.2020.129755
- Primary Citation of Related Structures:
6LJP, 6LJQ, 6LJR - PubMed Abstract:
The structure of human galectin-16 (Gal-16) has yet to be solved, and its function has remained elusive. X-ray crystallography was used to determine the atomic structures of Gal-16 and two of its mutants. The Gal-16 oligomer state was investigated by gel filtration, its hemagglutination activity was determined along with its ability to bind lactose using ITC. The cellular distribution of EGFP-tagged Gal-16 in various cell lines was also investigated, and the interaction between Gal-16 and c-Rel was assessed by pull-down studies, microscale thermophoresis and immunofluorescence. Unlike other galectins, Gal-16 lacks the ability to bind the β-galactoside lactose. Lactose binding could be regained by replacing an arginine (Arg55) with asparagine, as shown in the crystal structures of two lactose-loaded Gal-16 mutants (R55N and R55N/H57R). Gal-16 was also shown to be monomeric by gel filtration, as well as in crystal structures. Thus, this galectin could not induce erythrocyte agglutination. EGFP-tagged Gal-16 was found to be localized mostly in the nucleus of various cell types, and can interact with c-Rel, a member of NF-κB family. Gal-16 exists as a monomer and its ligand binding is significantly different from that of other prototype galectins, suggesting that it has a novel function(s). The interaction between Gal-16 and c-Rel indicates that Gal-16 may regulate signal transduction pathways via the c-Rel hub in B or T cells at the maternal-fetal interface. The present study lays the foundation for further studies into the cellular and physiological functions of Gal-16.
Organizational Affiliation:
Engineering Research Center of Glycoconjugates Ministry of Education, Jilin Provincial Key Laboratory of Chemistry and Biology of Changbai Mountain Natural Drugs, School of Life Sciences, Northeast Normal University, Changchun 130024, China; Jiangsu Key Laboratory of Brain Disease and Bioinformation, Research Center for Biochemistry and Molecular Biology, Xuzhou Medical University, Xuzhou 221004, China.