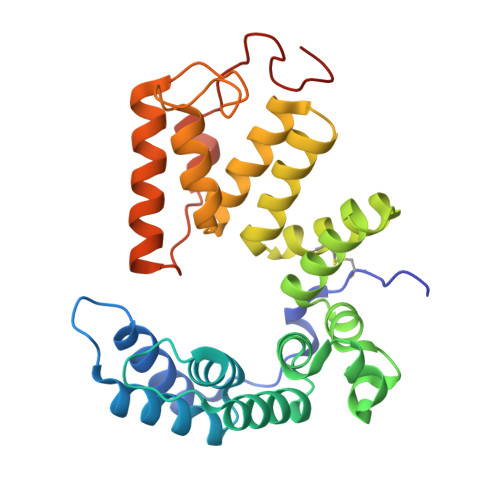
Characterization of the Pseudomonas aeruginosa T6SS PldB immunity proteins PA5086, PA5087 and PA5088 explains a novel stockpiling mechanism.
Wen, H., Geng, Z., Gao, Z., She, Z., Dong, Y.(2020) Acta Crystallogr F Struct Biol Commun 76: 222-227
- PubMed: 32356524
- DOI: https://doi.org/10.1107/S2053230X2000566X
- Primary Citation of Related Structures:
6LCH - PubMed Abstract:
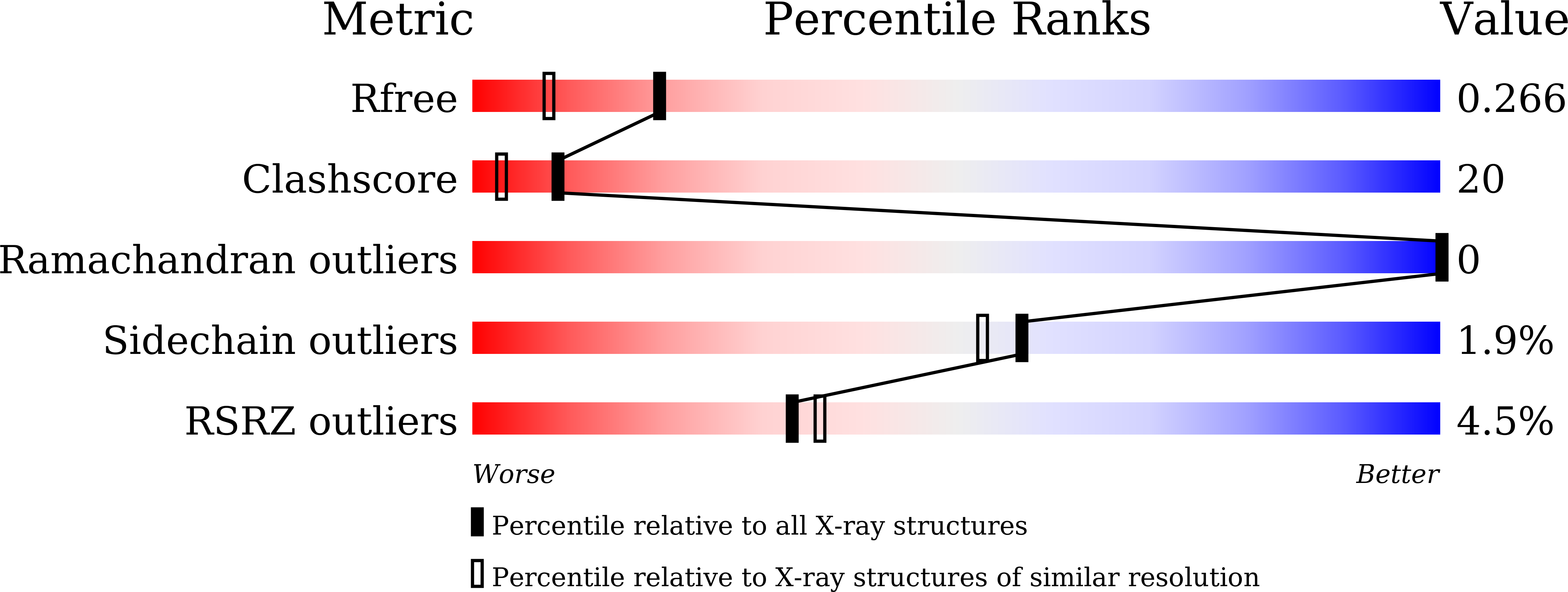
The bacterial type VI secretion system (T6SS) secretes many toxic effectors to gain advantage in interbacterial competition and for eukaryotic host infection. The cognate immunity proteins of these effectors protect bacteria from their own effectors. PldB is a T6SS trans-kingdom effector in Pseudomonas aeruginosa that can infect both prokaryotic and eukaryotic cells. Three proteins, PA5086, PA5087 and PA5088, are employed to suppress the toxicity of PldB-family proteins. The structures of PA5087 and PA5088 have previously been reported, but the identification of further distinctions between these immunity proteins is needed. Here, the crystal structure of PA5086 is reported at 1.90 Å resolution. A structural comparison of the three PldB immunity proteins showed vast divergences in their electrostatic potential surfaces. This interesting phenomenon provides an explanation of the stockpiling mechanism of T6SS immunity proteins.
Organizational Affiliation:
School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.