Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Chen, Z., Li, Z., Hu, X., Xie, F., Kuang, S., Zhan, B., Gao, W., Chen, X., Gao, S., Li, Y., Wang, Y., Qian, F., Ding, C., Gan, J., Ji, C., Xu, X., Zhou, Z., Huang, J., He, H.H., Li, J.(2020) Adv Sci (Weinh) 7: 2000532-2000532
- PubMed: 32714761
- DOI: https://doi.org/10.1002/advs.202000532
- Primary Citation of Related Structures:
6L5L, 6L5M, 6L5N, 6L5O - PubMed Abstract:
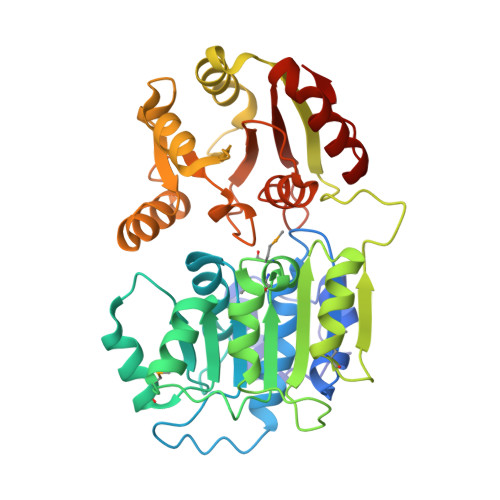
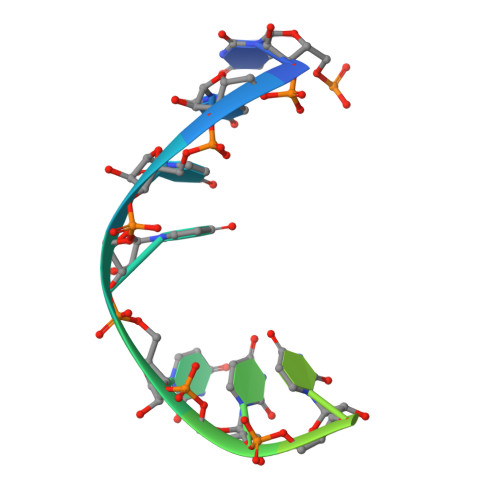
RNA helicase DDX21 plays vital roles in ribosomal RNA biogenesis, transcription, and the regulation of host innate immunity during virus infection. How DDX21 recognizes and unwinds RNA and how DDX21 interacts with virus remain poorly understood. Here, crystal structures of human DDX21 determined in three distinct states are reported, including the apo-state, the AMPPNP plus single-stranded RNA (ssRNA) bound pre-hydrolysis state, and the ADP-bound post-hydrolysis state, revealing an open to closed conformational change upon RNA binding and unwinding. The core of the RNA unwinding machinery of DDX21 includes one wedge helix, one sensor motif V and the DEVD box, which links the binding pockets of ATP and ssRNA. The mutant D339H/E340G dramatically increases RNA binding activity. Moreover, Hill coefficient analysis reveals that DDX21 unwinds double-stranded RNA (dsRNA) in a cooperative manner. Besides, the nonstructural (NS1) protein of influenza A inhibits the ATPase and unwinding activity of DDX21 via small RNAs, which cooperatively assemble with DDX21 and NS1. The structures illustrate the dynamic process of ATP hydrolysis and RNA unwinding for RNA helicases, and the RNA modulated interaction between NS1 and DDX21 generates a fresh perspective toward the virus-host interface. It would benefit in developing therapeutics to combat the influenza virus infection.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering Department of Neurology School of Life Sciences and Huashan Hospital Collaborative Innovation Center of Genetics and Development Engineering Research Center of Gene Technology of MOE Shanghai Engineering Research Center of Industrial Microorganisms Fudan University Shanghai 200438 China.