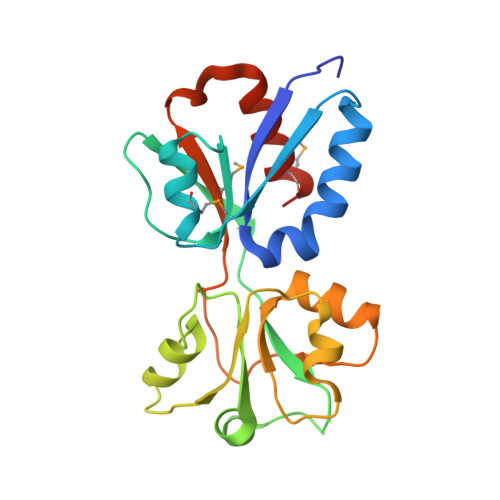
Crystal Structure of the Regulatory Domain of MexT, a Transcriptional Activator of the MexEFOprN Efflux Pump inPseudomonas aeruginosa.
Kim, S., Kim, S.H., Ahn, J., Jo, I., Lee, Z.W., Choi, S.H., Ha, N.(2019) Mol Cells 42: 850-857
- PubMed: 31722511
- DOI: https://doi.org/10.14348/molcells.2019.0168
- Primary Citation of Related Structures:
6L33 - PubMed Abstract:
The Gram-negative opportunistic pathogen, Pseudomonas aeruginosa , has multiple multidrug efflux pumps. MexT, a LysR-type transcriptional regulator, functions as a transcriptional activator of the MexEF-OprN efflux system. MexT consists of an N-terminal DNA-binding domain and a C-terminal regulatory domain (RD). Little is known regarding MexT ligands and its mechanism of activation. We elucidated the crystal structure of the MexT RD at 2.0 Å resolution. The structure comprised two protomer chains in a dimeric arrangement. MexT possessed an arginine-rich region and a hydrophobic patch lined by a variable loop, both of which are putative ligand-binding sites. The three-dimensional structure of MexT provided clues to the interacting ligand structure. A DNase I footprinting assay of full-length MexT identified two MexT-binding sequence in the mexEF oprN promoter. Our findings enhance the understanding of the regulation of MexT-dependent activation of efflux pumps.
Organizational Affiliation:
Department of Agricultural Biotechnology, Center for Food Safety and Toxicology, Center for Food and Bioconvergence, and Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul 08826, Korea.