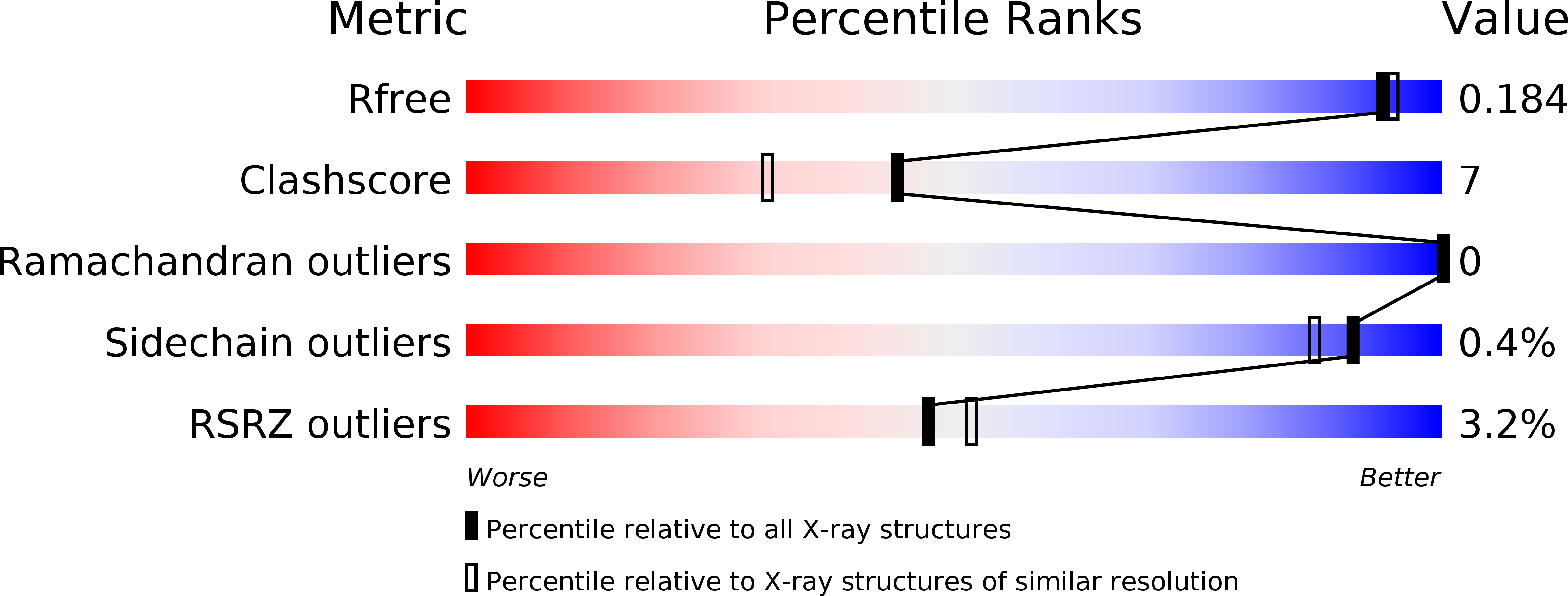
Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Cui, S., Wang, Y., Wang, Y., Tang, X., Ren, X., Zhang, L., Xu, Y., Zhang, Z., Zhang, Z.M., Lu, X., Ding, K.(2019) Eur J Med Chem 179: 470-482
- PubMed: 31271959
- DOI: https://doi.org/10.1016/j.ejmech.2019.06.064
- Primary Citation of Related Structures:
6KZC, 6KZD - PubMed Abstract:
A series of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides was designed and synthesized as new tropomyosin receptor kinases (Trks) inhibitors by utilizing a structure-guided optimization strategy. One of the most potent compounds 9o suppressed TrkA/B/C with IC 50 values of 2.65, 10.47 and 2.95 nM, respectively. The compound dose-dependently inhibited brain-derived neurotrophic factor (BDNF)-mediated TrkB activation and suppressed migration and invasion of SH-SY5Y-TrkB neuroblastoma cells expressing high level of TrkB. Inhibitor 9o also inhibited the proliferation of SH-SY5Y-TrkB cells with an IC 50 value of 58 nM, which was comparable to that of an US FDA recently approved drug LOXO-101. Compound 9o may serve as a new lead compound for further anti-cancer drug discovery.
Organizational Affiliation:
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, No. 190 Kaiyuan Avenue, Guangzhou, 510530, China; University of Chinese Academy of Sciences, No. 19 Yuquan Road, Beijing, 100049, China.