Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
Dong, F., Xu, F., Chen, X.L., Li, P.Y., Li, C.Y., Li, F.C., Chen, Y., Wang, P., Zhang, Y.Z.(2019) J Mol Biol 431: 4897-4909
- PubMed: 31682837
- DOI: https://doi.org/10.1016/j.jmb.2019.10.023
- Primary Citation of Related Structures:
6KCV, 6KCW, 6KZK - PubMed Abstract:
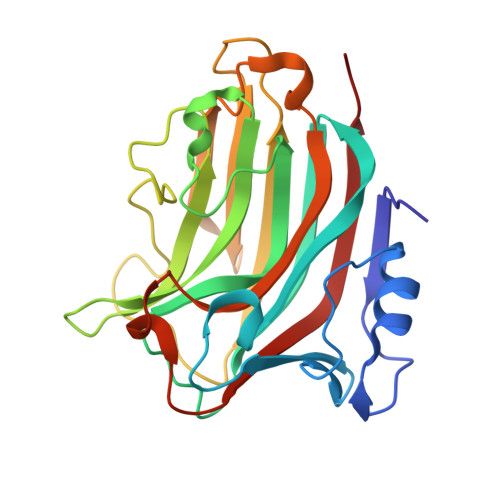
Alginate lyases, which are important in both basic and applied sciences, fall into ten polysaccharide lyase (PL) families. PL36 is a newly established family that includes 39 bacterial sequences and one eukaryotic sequence. Till now, the structures or catalytic mechanisms of PL36 alginate lyases have yet to be revealed. Here, we characterized a novel PL36 alginate lyase, Aly36B, from Chitinophaga sp. MD30. Aly36B is a polymannuronate specific endolytic alginate lyase. To probe the catalytic mechanism of Aly36B, the structures of wild-type Aly36B and its mutants (K143A/Y185A in complex with alginate tetrasaccharide and K143A/M171A with trisaccharide) were solved. The overall structure of Aly36B belongs to the β-jelly roll scaffold, adopting a typical β-sandwich fold. Aly36B contains a Ca 2+ , which is far away from the active center and plays an important role in stabilizing the structure of Aly36B. Based on structural and mutational analyses, the catalytic mechanism of Aly36B for alginate degradation was explained. During catalysis, Arg 169 , Tyr 185 , and Tyr 187 are responsible for neutralizing the negative charge of the substrate, and Lys 143 acts as both the catalytic base and the catalytic acid, which represents a new kind of catalytic mechanism of alginate lyases. Sequence alignment shows that these four residues involved in catalysis are highly conserved in all PL36 sequences, suggesting that PL36 alginate lyases may adopt a similar catalytic mechanism. Taken together, this study reveals the molecular structure and catalytic mechanism of a PL36 alginate lyase, broadening our knowledge on alginate lyases and facilitating future biotechnological applications of PL36 alginate lyases.
Organizational Affiliation:
State Key Laboratory of Microbial Technology, Marine Biotechnology Research Center, Shandong University, Qingdao, 266237, China.