Structural Characterization ofArabidopsis thalianaNAP1-Related Protein 2 (AtNRP2) and Comparison with its Homolog AtNRP1.
Kumar, A., Kumar Singh, A., Chandrakant Bobde, R., Vasudevan, D.(2019) Molecules 24
- PubMed: 31213016
- DOI: https://doi.org/10.3390/molecules24122258
- Primary Citation of Related Structures:
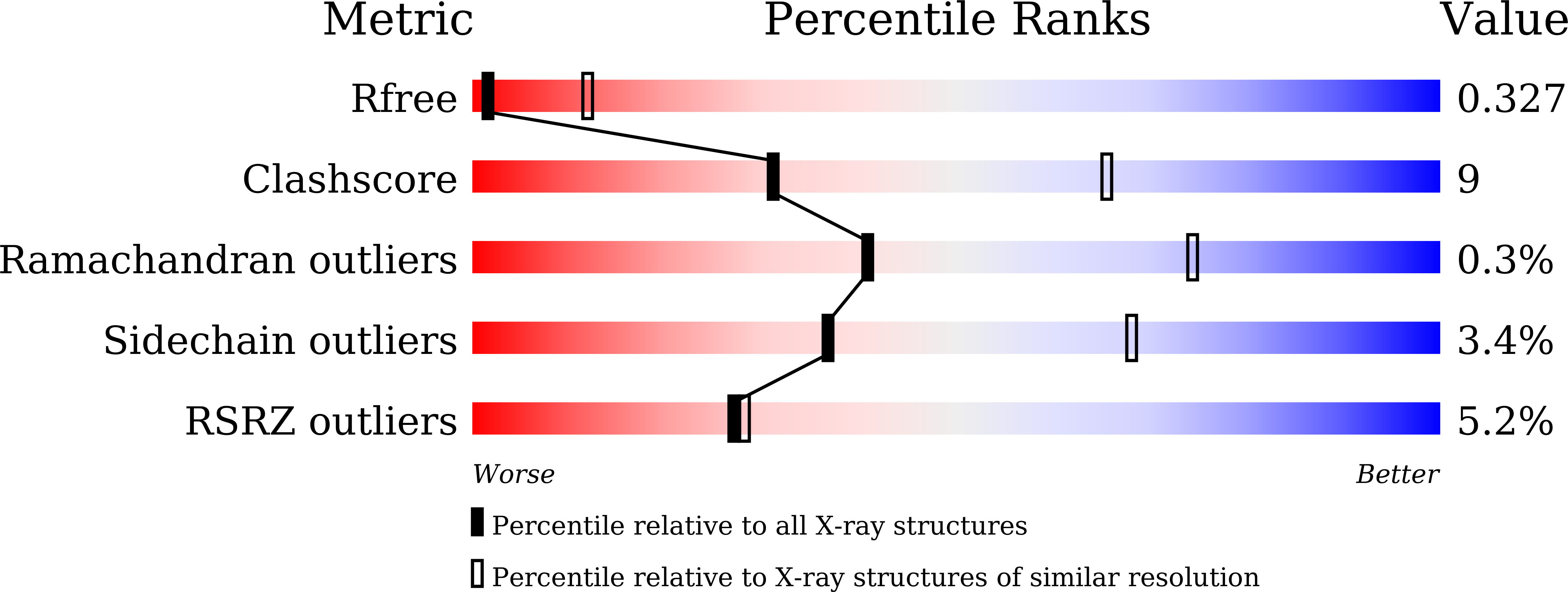
6JQV - PubMed Abstract:
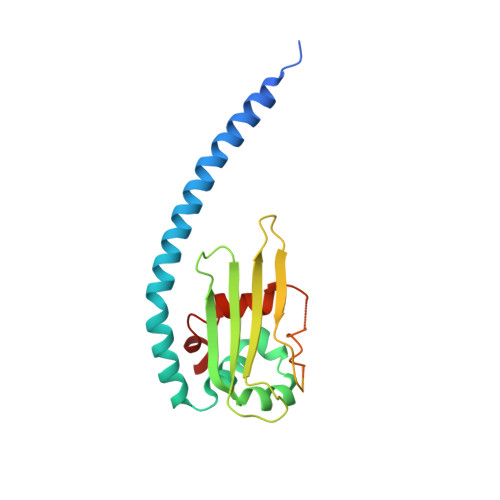
Nucleosome Assembly Protein (NAP) is a highly conserved family of histone chaperones present in yeast, animals, and plants. Unlike other organisms, plants possess an additional class of proteins in its NAP family, known as the NAP1-related proteins or NRP. Arabidopsis thaliana possesses two NRP isoforms, namely AtNRP1 and AtNRP2, that share 87% sequence identity. Both AtNRP1 and AtNRP2 get expressed in all the plant tissues. Most works in the past, including structural studies, have focused on AtNRP1. We wanted to do a comparative study of the two proteins to find why the plant would have two very similar proteins and whether there is any difference between the two for their structure and function as histone chaperones. Here we report the crystal structure of AtNRP2 and a comparative analysis of its structural architecture with other NAP family proteins. The crystal structure of AtNRP2 shows it to be a homodimer, with its fold similar to that of other structurally characterized NAP family proteins. Although AtNRP1 and AtNRP2 have a similar fold, upon structural superposition, we find an offset in the dimerization helix of the two proteins. We evaluated the stability, oligomerization status, and histone chaperoning properties of the two proteins, for a comparison. The thermal melting experiments suggest that AtNRP2 is more stable than AtNRP1 at higher temperatures. In addition, electrophoretic mobility shift assay and isothermal titration calorimetry experiments suggest histone binding ability of AtNRP2 is higher than that of AtNRP1. Overall, these results provide insights about the specific function and relevance of AtNRP2 in plants through structural and biophysical studies.
Organizational Affiliation:
Institute of Life Sciences, Bhubaneswar 751023, Odisha, India. mail2ashish.bt@gmail.com.