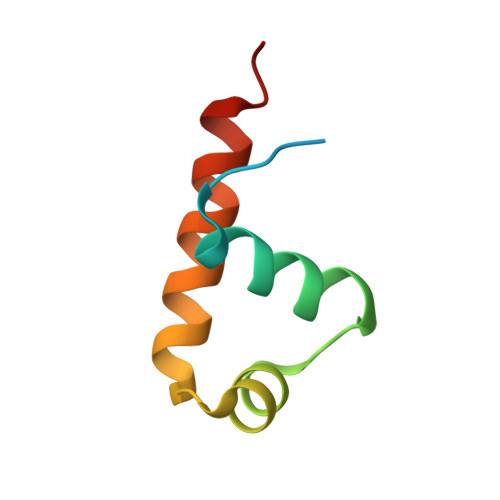
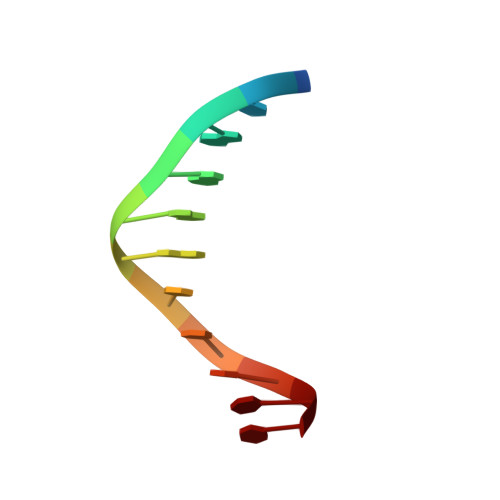
Structural analysis of the recognition of the -35 promoter element by SigW from Bacillus subtilis.
Kwon, E., Devkota, S.R., Pathak, D., Dahal, P., Kim, D.Y.(2019) PLoS One 14: e0221666-e0221666
- PubMed: 31461489
- DOI: https://doi.org/10.1371/journal.pone.0221666
- Primary Citation of Related Structures:
6JHE - PubMed Abstract:
Sigma factors are key proteins that mediate the recruitment of RNA polymerase to the promoter regions of genes, for the initiation of bacterial transcription. Multiple sigma factors in a bacterium selectively recognize their cognate promoter sequences, thereby inducing the expression of their own regulons. In this paper, we report the crystal structure of the σ4 domain of Bacillus subtilis SigW bound to the -35 promoter element. Purine-specific hydrogen bonds of the -35 promoter element with the recognition helix α9 of the σ4 domain occurs at three nucleotides of the consensus sequence (G-35, A-34, and G'-31 in G-35A-34A-33A-32C-31C-30T-29). The hydrogen bonds of the backbone with the α7 and α8 of the σ4 domain occurs at G'-30. These results elucidate the structural basis of the selective recognition of the promoter by SigW. In addition, comparison of SigW structures complexed with the -35 promoter element or with anti-sigma RsiW reveals that DNA recognition and anti-sigma factor binding of SigW are mutually exclusive.
Organizational Affiliation:
College of Pharmacy, Yeungnam University, Gyeongsan, Gyeongbuk, South Korea.