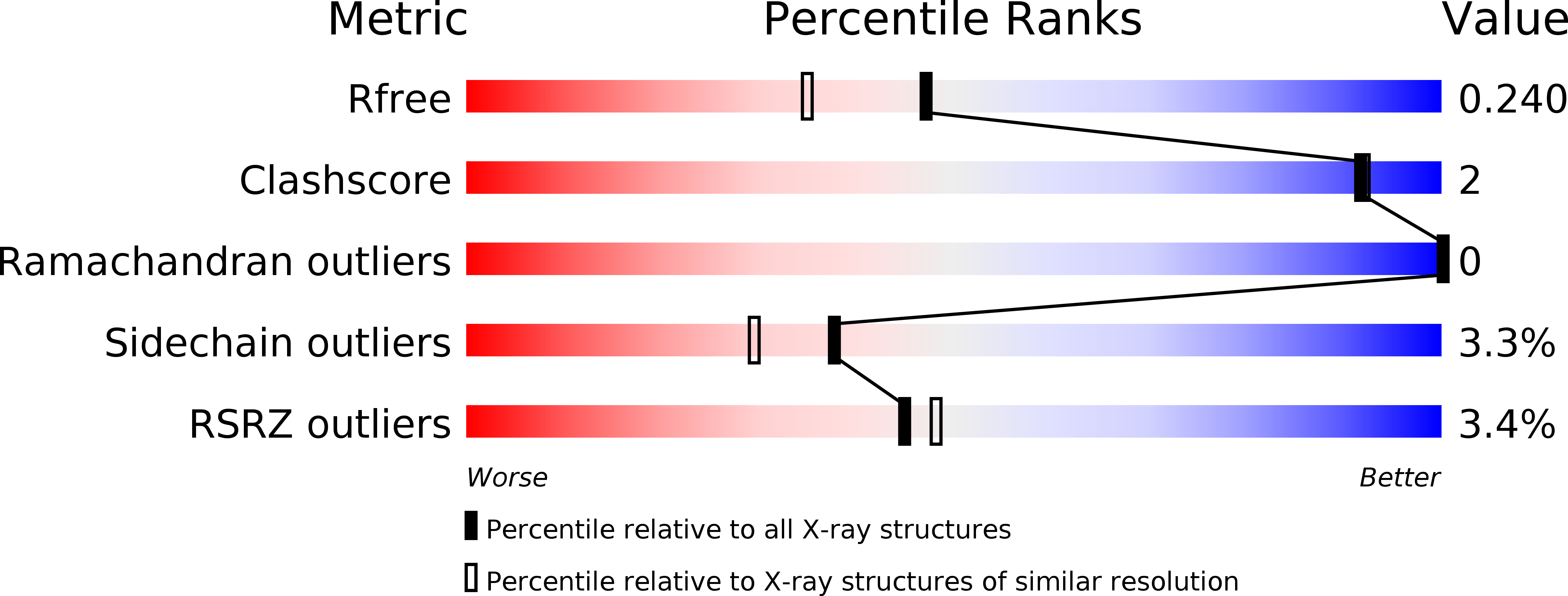
Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Miyake, Y., Keusch, J.J., Decamps, L., Ho-Xuan, H., Iketani, S., Gut, H., Kutay, U., Helenius, A., Yamauchi, Y.(2019) Nat Microbiol 4: 578-586
- PubMed: 30692667
- DOI: https://doi.org/10.1038/s41564-018-0332-2
- Primary Citation of Related Structures:
6I3H - PubMed Abstract:
Influenza A virus is a pathogen of great medical impact. To develop novel antiviral strategies, it is essential to understand the molecular aspects of virus-host cell interactions in detail. During entry, the viral ribonucleoproteins (vRNPs) that carry the RNA genome must be released from the incoming particle before they can enter the nucleus for replication. The uncoating process is facilitated by histone deacetylase 6 (ref. 1 ). However, the precise mechanism of shell opening and vRNP debundling is unknown. Here, we show that transportin 1, a member of the importin-β family proteins, binds to a PY-NLS 2 sequence motif close to the amino terminus of matrix protein (M1) exposed during acid priming of the viral core. It promotes the removal of M1 and induces disassembly of vRNP bundles. Next, the vRNPs interact with importin-α/β and enter the nucleus. Thus, influenza A virus uses dual importin-βs for distinct steps in host cell entry.
Organizational Affiliation:
School of Cellular and Molecular Medicine, Faculty of Life Sciences, University of Bristol, Bristol, UK.