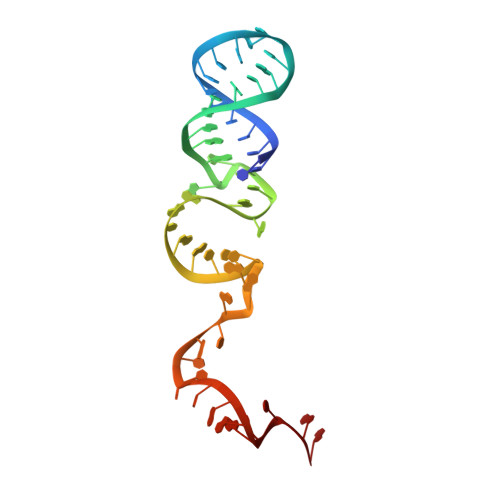Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Zhang, C., Konermann, S., Brideau, N.J., Lotfy, P., Wu, X., Novick, S.J., Strutzenberg, T., Griffin, P.R., Hsu, P.D., Lyumkis, D.(2018) Cell 175: 212-223.e17
- PubMed: 30241607
- DOI: https://doi.org/10.1016/j.cell.2018.09.001
- Primary Citation of Related Structures:
6E9E, 6E9F - PubMed Abstract:
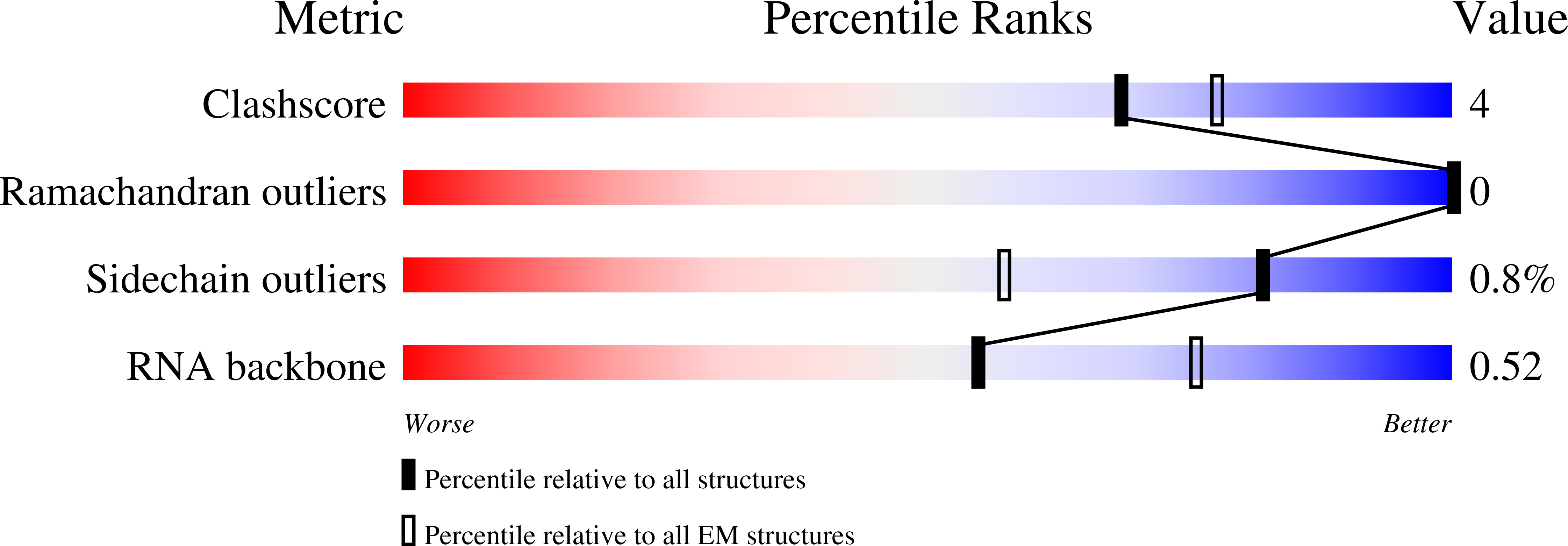
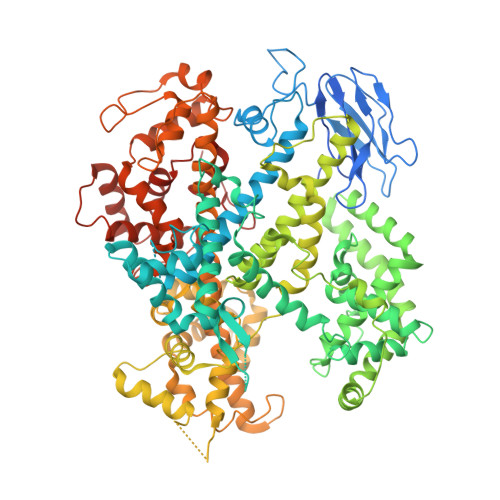
CRISPR-Cas endonucleases directed against foreign nucleic acids mediate prokaryotic adaptive immunity and have been tailored for broad genetic engineering applications. Type VI-D CRISPR systems contain the smallest known family of single effector Cas enzymes, and their signature Cas13d ribonuclease employs guide RNAs to cleave matching target RNAs. To understand the molecular basis for Cas13d function and explain its compact molecular architecture, we resolved cryoelectron microscopy structures of Cas13d-guide RNA binary complex and Cas13d-guide-target RNA ternary complex to 3.4 and 3.3 Å resolution, respectively. Furthermore, a 6.5 Å reconstruction of apo Cas13d combined with hydrogen-deuterium exchange revealed conformational dynamics that have implications for RNA scanning. These structures, together with biochemical and cellular characterization, provide insights into its RNA-guided, RNA-targeting mechanism and delineate a blueprint for the rational design of improved transcriptome engineering technologies.
Organizational Affiliation:
Laboratory of Genetics, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA; Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA.