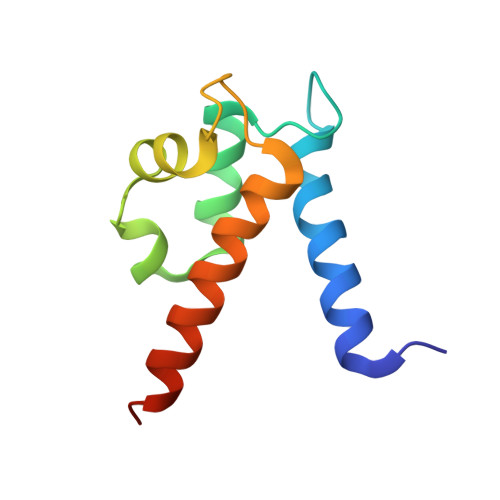
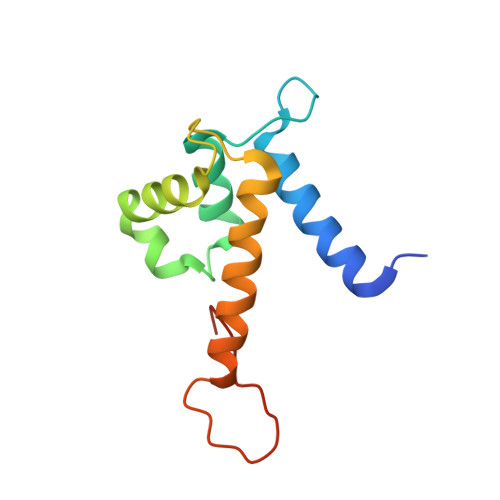
Biophysical Examination of the Calcium-Modulated Nickel-Binding Properties of Human Calprotectin Reveals Conformational Change in the EF-Hand Domains and His3Asp Site.
Nakashige, T.G., Bowman, S.E.J., Zygiel, E.M., Drennan, C.L., Nolan, E.M.(2018) Biochemistry 57: 4155-4164
- PubMed: 29890074
- DOI: https://doi.org/10.1021/acs.biochem.8b00415
- Primary Citation of Related Structures:
6DS2 - PubMed Abstract:
Calprotectin (CP, S100A8/S100A9 oligomer, MRP-8/MRP-14 oligomer) is a host-defense protein that sequesters nutrient transition metals from microbes. Each S100A8/S100A9 heterodimer contains four EF-hand domains and two transition-metal-binding sites. We investigate the effect of Ca(II) ions on the structure and Ni(II)-binding properties of human CP. By employing energy dispersive X-ray (EDX) spectroscopy, we evaluate the metal content of Ni(II)-bound CP-Ser [oligomer of S100A8(C42S) and S100A9(C3S)] crystals obtained in the absence and presence of Ca(II). We present a 2.1 Å resolution crystal structure of Ni(II)-bound CP-Ser and compare this structure to a reported Ni(II)- and Ca(II)-bound CP-Ser structure [Nakashige, T. G., et al. (2017) J. Am. Chem. Soc. 139, 8828-8836]. This analysis reveals conformational changes associated with coordination of Ca(II) to the EF-hands of S100A9 and that Ca(II) binding affects the coordination number and geometry of the Ni(II) ion bound to the His 3 Asp site. In contrast, negligible differences are observed for the Ni(II)-His 6 site in the absence and presence of Ca(II). Biochemical studies show that, whereas the His 6 site has a thermodynamic preference for Ni(II) over Zn(II), the His 3 Asp site selects for Zn(II) over Ni(II), and relatively rapid metal exchange occurs at this site. These observations inform the working model for how CP withholds nutrient metals in the extracellular space.