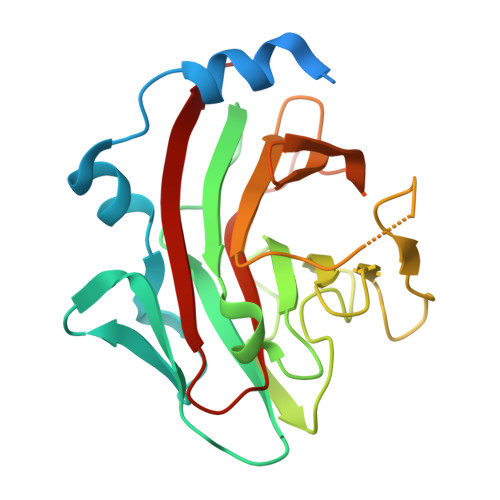
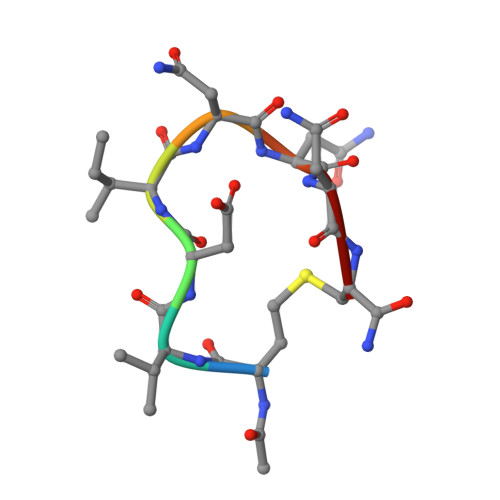
A Cyclic Peptide Inhibitor of the iNOS-SPSB Protein-Protein Interaction as a Potential Anti-Infective Agent.
Sadek, M.M., Barlow, N., Leung, E.W.W., Williams-Noonan, B.J., Yap, B.K., Shariff, F.M., Caradoc-Davies, T.T., Nicholson, S.E., Chalmers, D.K., Thompson, P.E., Law, R.H.P., Norton, R.S.(2018) ACS Chem Biol 13: 2930-2938
- PubMed: 30226743
- DOI: https://doi.org/10.1021/acschembio.8b00561
- Primary Citation of Related Structures:
6DN5, 6DN6, 6DN7, 6DN8 - PubMed Abstract:
SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4 interact with inducible nitric oxide synthase (iNOS), causing the iNOS to be polyubiquitinated and targeted for degradation. Inhibition of this interaction increases iNOS levels, and consequently cellular nitric oxide (NO) concentrations, and has been proposed as a potential strategy for killing intracellular pathogens. We previously described two DINNN-containing cyclic peptides (CP1 and CP2) as potent inhibitors of the murine SPSB-iNOS interaction. In this study, we report the crystal structures of human SPSB4 bound to CP1 and CP2 and human SPSB2 bound to CP2. We then used these structures to design a new inhibitor in which an intramolecular hydrogen bond was replaced with a hydrocarbon linkage to form a smaller macrocycle while maintaining the bound geometry of CP2 observed in the crystal structures. This resulting pentapeptide SPSB-iNOS inhibitor (CP3) has a reduced macrocycle ring size, fewer nonbinding residues, and includes additional conformational constraints. CP3 has a greater affinity for SBSB2 ( K D = 7 nM as determined by surface plasmon resonance) and strongly inhibits the SPSB2-iNOS interaction in macrophage cell lysates. We have also determined the crystal structure of CP3 in complex with human SPSB2, which reveals the structural basis for the increased potency of CP3 and validates the original design.
Organizational Affiliation:
Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences , Monash University , Parkville Victoria 3052 , Australia.