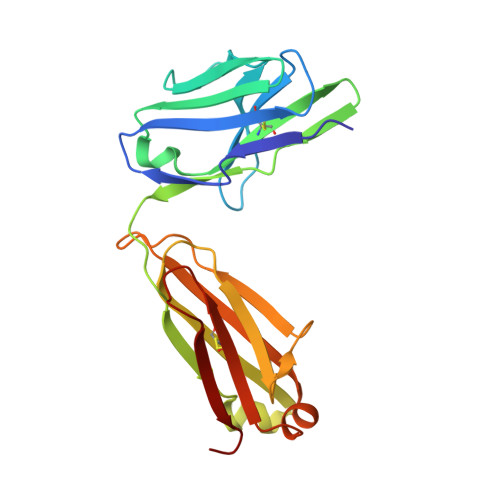
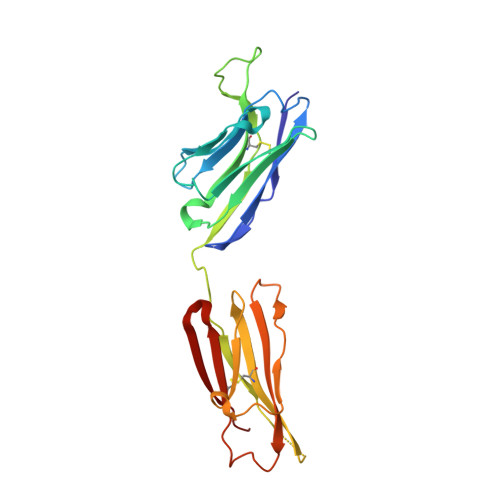
Oligomannose Glycopeptide Conjugates Elicit Antibodies Targeting the Glycan Core Rather than Its Extremities.
Nguyen, D.N., Xu, B., Stanfield, R.L., Bailey, J.K., Horiya, S., Temme, J.S., Leon, D.R., LaBranche, C.C., Montefiori, D.C., Costello, C.E., Wilson, I.A., Krauss, I.J.(2019) ACS Cent Sci 5: 237-249
- PubMed: 30834312
- DOI: https://doi.org/10.1021/acscentsci.8b00588
- Primary Citation of Related Structures:
6CXG, 6CXL - PubMed Abstract:
Up to ∼20% of HIV-infected individuals eventually develop broadly neutralizing antibodies (bnAbs), and many of these antibodies (∼40%) target a region of dense high-mannose glycosylation on gp120 of the HIV envelope protein, known as the "high-mannose patch" (HMP). Thus, there have been numerous attempts to develop glycoconjugate vaccine immunogens that structurally mimic the HMP and might elicit bnAbs targeting this conserved neutralization epitope. Herein, we report on the immunogenicity of glycopeptides, designed by in vitro selection, that bind tightly to anti-HMP antibody 2G12. By analyzing the fine carbohydrate specificity of rabbit antibodies elicited by these immunogens, we found that they differ from some natural human bnAbs, such as 2G12 and PGT128, in that they bind primarily to the core structures within the glycan, rather than to the Manα1 → 2Man termini (2G12) or to the whole glycan (PGT128). Antibody specificity for the glycan core may result from extensive serum mannosidase trimming of the immunogen in the vaccinated animals. This finding has broad implications for vaccine design aiming to target glycan-dependent HIV neutralizing antibodies.
Organizational Affiliation:
Department of Chemistry, Brandeis University, Waltham, Massachusetts 02454-9110, United States.