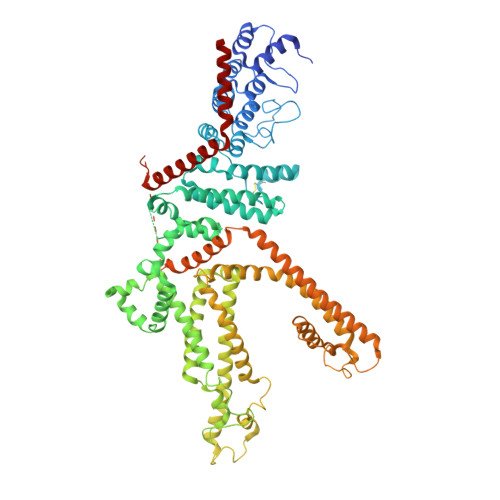
Structure of the human lipid-gated cation channel TRPC3.
Fan, C., Choi, W., Sun, W., Du, J., Lu, W.(2018) Elife 7
- PubMed: 29726814
- DOI: https://doi.org/10.7554/eLife.36852
- Primary Citation of Related Structures:
6CUD - PubMed Abstract:
The TRPC channels are crucially involved in store-operated calcium entry and calcium homeostasis, and they are implicated in human diseases such as neurodegenerative disease, cardiac hypertrophy, and spinocerebellar ataxia. We present a structure of the full-length human TRPC3, a lipid-gated TRPC member, in a lipid-occupied, closed state at 3.3 Angstrom. TRPC3 has four elbow-like membrane reentrant helices prior to the first transmembrane helix. The TRP helix is perpendicular to, and thus disengaged from, the pore-lining S6, suggesting a different gating mechanism from other TRP subfamily channels. The third transmembrane helix S3 is remarkably long, shaping a unique transmembrane domain, and constituting an extracellular domain that may serve as a sensor of external stimuli. We identified two lipid-binding sites, one being sandwiched between the pre-S1 elbow and the S4-S5 linker, and the other being close to the ion-conducting pore, where the conserved LWF motif of the TRPC family is located.
Organizational Affiliation:
Van Andel Institute, Grand Rapids, United States.