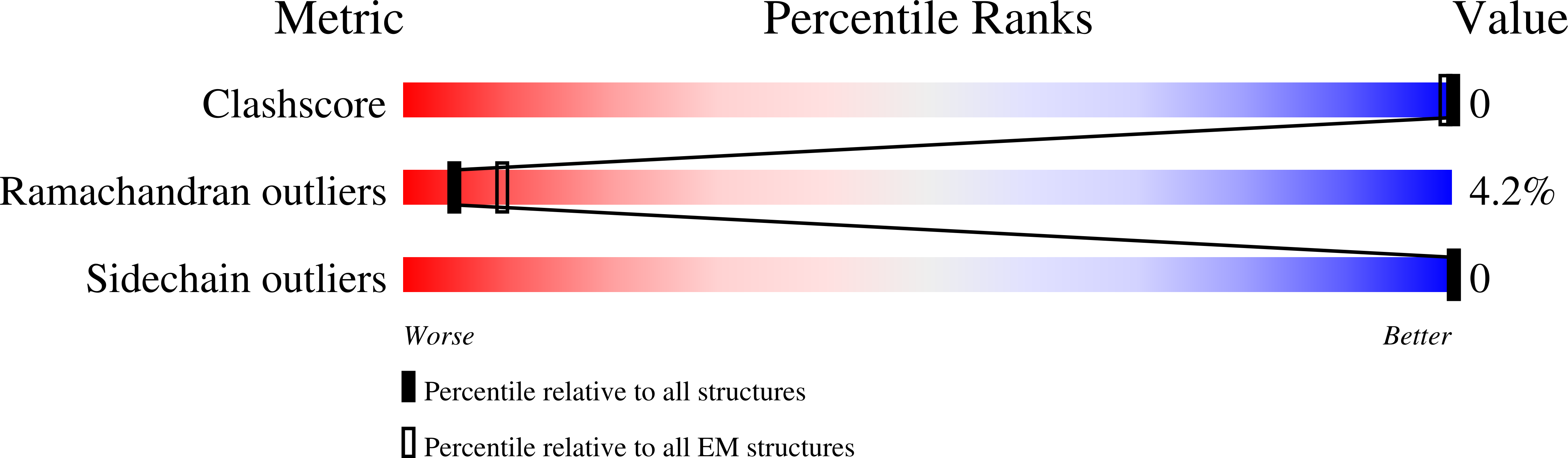Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80 alpha and SaPI1 Capsids.
Kizziah, J.L., Manning, K.A., Dearborn, A.D., Wall, E.A., Klenow, L., Hill, R.L.L., Spilman, M.S., Stagg, S.M., Christie, G.E., Dokland, T.(2017) Viruses 9
- PubMed: 29258203
- DOI: https://doi.org/10.3390/v9120384
- Primary Citation of Related Structures:
6C21, 6C22 - PubMed Abstract:
In the tailed bacteriophages, DNA is packaged into spherical procapsids, leading to expansion into angular, thin-walled mature capsids. In many cases, this maturation is accompanied by cleavage of the major capsid protein (CP) and other capsid-associated proteins, including the scaffolding protein (SP) that serves as a chaperone for the assembly process. Staphylococcus aureus bacteriophage 80α is capable of high frequency mobilization of mobile genetic elements called S. aureus pathogenicity islands (SaPIs), such as SaPI1. SaPI1 redirects the assembly pathway of 80α to form capsids that are smaller than those normally made by the phage alone. Both CP and SP of 80α are N-terminally processed by a host-encoded protease, Prp. We have analyzed phage mutants that express pre-cleaved or uncleavable versions of CP or SP, and show that the N-terminal sequence in SP is absolutely required for assembly, but does not need to be cleaved in order to produce viable capsids. Mutants with pre-cleaved or uncleavable CP display normal viability. We have used cryo-EM to solve the structures of mature capsids from an 80α mutant expressing uncleavable CP, and from wildtype SaPI1. Comparisons with structures of 80α and SaPI1 procapsids show that capsid maturation involves major conformational changes in CP, consistent with a release of the CP N-arm by SP. The hexamers reorganize during maturation to accommodate the different environments in the 80α and SaPI1 capsids.
Organizational Affiliation:
Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294, USA. kizziah4@uab.edu.