Structure-Based Design of Novel EphA2 Agonistic Agents with Nanomolar Affinity in Vitro and in Cell.
Gambini, L., Salem, A.F., Udompholkul, P., Tan, X.F., Baggio, C., Shah, N., Aronson, A., Song, J., Pellecchia, M.(2018) ACS Chem Biol 13: 2633-2644
- PubMed: 30110533
- DOI: https://doi.org/10.1021/acschembio.8b00556
- Primary Citation of Related Structures:
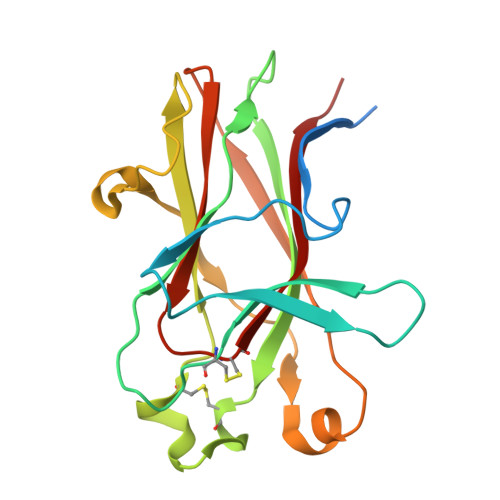
6B9L - PubMed Abstract:
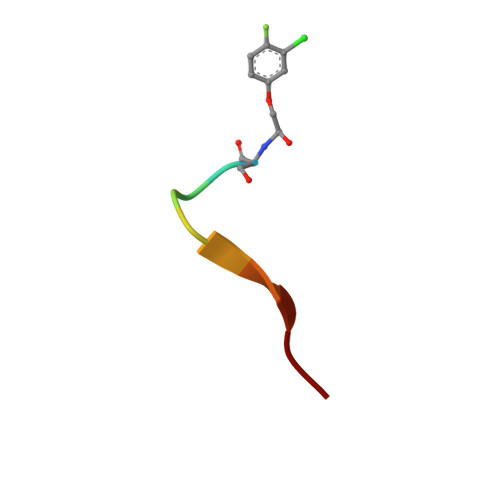
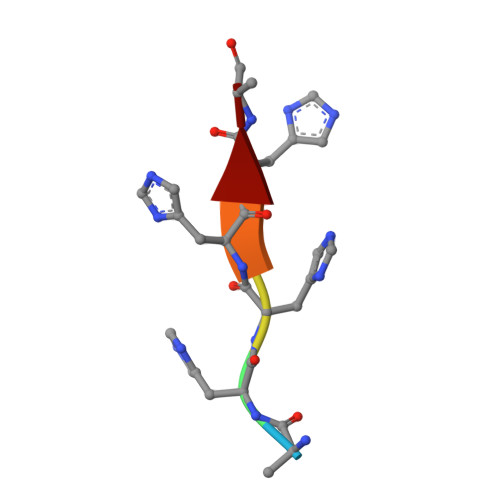
EphA2 overexpression is invariably associated with poor prognosis and development of aggressive metastatic cancers in pancreatic, prostate, lung, ovarian, and breast cancers and melanoma. Recent efforts from our laboratories identified a number of agonistic peptides targeting the ligand-binding domain of the EphA2 receptor. The individual agents, however, were still relatively weak in affinities (micromolar range) that precluded detailed structural studies on the mode of action. Using a systematic optimization of the 12-mer peptide mimetic 123B9, we were able to first derive an agent that displayed a submicromolar affinity for the receptor. This agent enabled cocrystallization with the EphA2 ligand-binding domain providing for the first time the structural basis for their agonistic mechanism of action. In addition, the atomic coordinates of the complex enabled rapid iterations of structure-based optimizations that resulted in a novel agonistic agent, named 135H11, with a nanomolar affinity for the receptor, as demonstrated by in vitro binding assays (isothermal titration calorimetry measurements), and a biochemical displacement assay. As we have recently demonstrated, the cellular activity of these agents is further increased by synthesizing dimeric versions of the compounds. Hence, we report that a dimeric version of 135H11 is extremely effective at low nanomolar concentrations to induce cellular receptor activation, internalization, and inhibition of cell migration in a pancreatic cancer cell line. Given the pivotal role of EphA2 in tumor growth, angiogenesis, drug resistance, and metastasis, these agents, and the associated structural studies, provide significant advancements in the field for the development of novel EphA2-targeting therapeutics or diagnostics.
Organizational Affiliation:
Division of Biomedical Sciences, School of Medicine , University of California Riverside , 900 University Avenue , Riverside , California 92521 , United States.