Slow luminescence kinetics of semi-synthetic aequorin: expression, purification and structure determination of cf3-aequorin.
Inouye, S., Tomabechi, Y., Hosoya, T., Sekine, S.I., Shirouzu, M.(2018) J Biochem 164: 247-255
- PubMed: 29796619
- DOI: https://doi.org/10.1093/jb/mvy049
- Primary Citation of Related Structures:
5ZAB - PubMed Abstract:
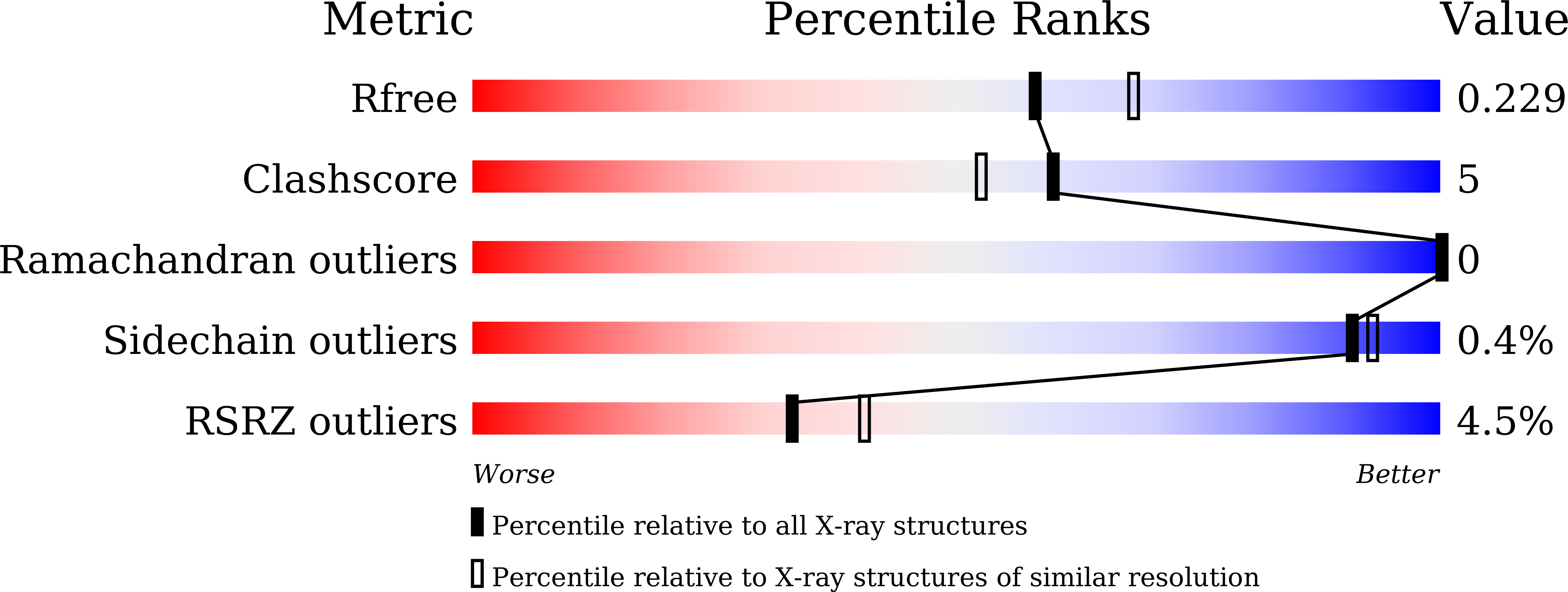
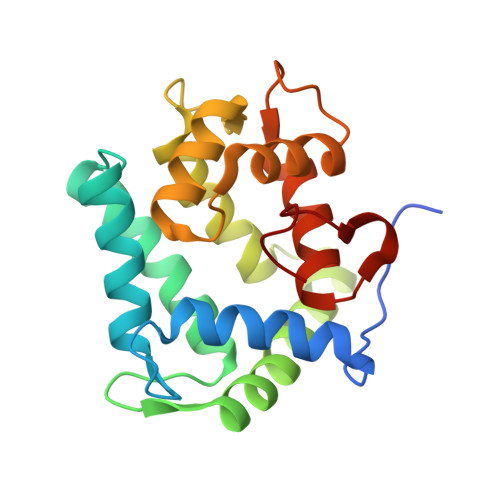
cf3-Aequorin is one of the semi-synthetic aequorins that was produced by replacing 2-peroxycoelenterazine (CTZ-OOH) in native aequorin with a 2-peroxycoelenterazine analog, and it was prepared using the C2-modified trifluoromethyl analog of coelenterazine (cf3-CTZ) and the histidine-tagged apoaequorin expressed in Escherichia coli cells. The purified cf3-aequorin showed a slow luminescence pattern with half-decay time of maximum intensities of luminescence of 5.0 s. This is much longer than that of 0.9 s for native aequorin, and its luminescence capacity was estimated to be 72.8% of that of native aequorin. The crystal structure of cf3-aequorin was determined at 2.15 Å resolution. The light source of 2-peroxytrifluoromethylcoelenterazine (cf3-CTZ-OOH) was stabilized by the hydrogen-bonding interactions at the C2-peroxy moiety and the p-hydroxy moiety at the C6-phenyl group. In native aequorin, three water molecules contribute to stabilizing CTZ-OOH through hydrogen bonds. However, cf3-aequorin only contained one water molecule, and the trifluoromethyl moiety at the C2-benzyl group of cf3-CTZ-OOH interacted with the protein by van der Waals interactions. The slow luminescence kinetics of cf3-aequorin could be explained by slow conformational changes due to the bulkiness of the trifluoromethyl group, which might hinder the smooth cleavage of hydrogen bonds at the C2-peroxy moiety after the binding of Ca2+ to cf3-aequorin.
Organizational Affiliation:
Yokohama Research Center, JNC. Co., 5-1 Okawa, Kanazawa-ku, Yokohama, Japan.