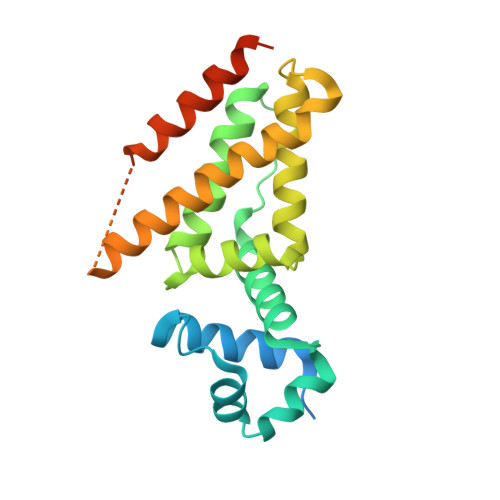
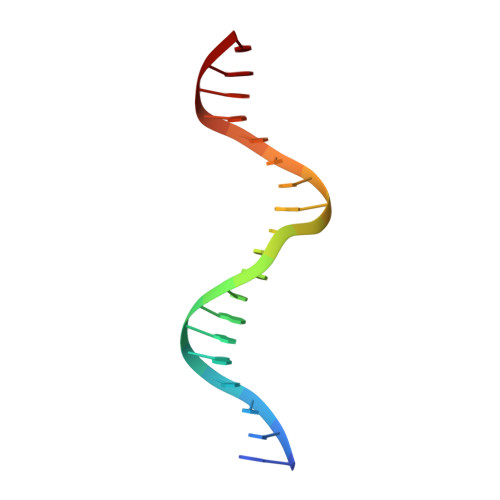
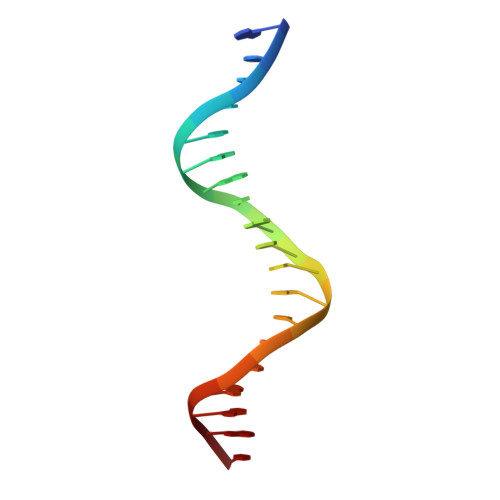
Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Yan, L., Tang, Q., Guan, Z., Pei, K., Zou, T., He, J.(2018) Biochim Biophys Acta Gen Subj 1862: 1843-1851
- PubMed: 29852200
- DOI: https://doi.org/10.1016/j.bbagen.2018.05.015
- Primary Citation of Related Structures:
5YEJ, 5YEK - PubMed Abstract:
Biotin is an essential cofactor in living organisms. The TetR family transcriptional regulator (TFTR) BioQ is the main regulator of biotin synthesis in Mycobacterium smegmatis. BioQ represses the expression of its target genes by binding to a conserved palindromic DNA sequence (the BioQ operator). However, the mechanism by which BioQ recognizes this DNA element has not yet been fully elucidated.
Organizational Affiliation:
College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.