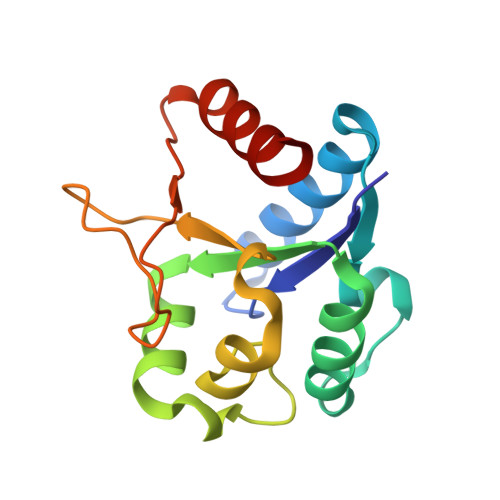
Structure of a prokaryotic SEFIR domain reveals two novel SEFIR-SEFIR interaction modes.
Yang, H., Zhu, Y., Chen, X., Li, X., Ye, S., Zhang, R.(2018) J Struct Biol 203: 81-89
- PubMed: 29549035
- DOI: https://doi.org/10.1016/j.jsb.2018.03.005
- Primary Citation of Related Structures:
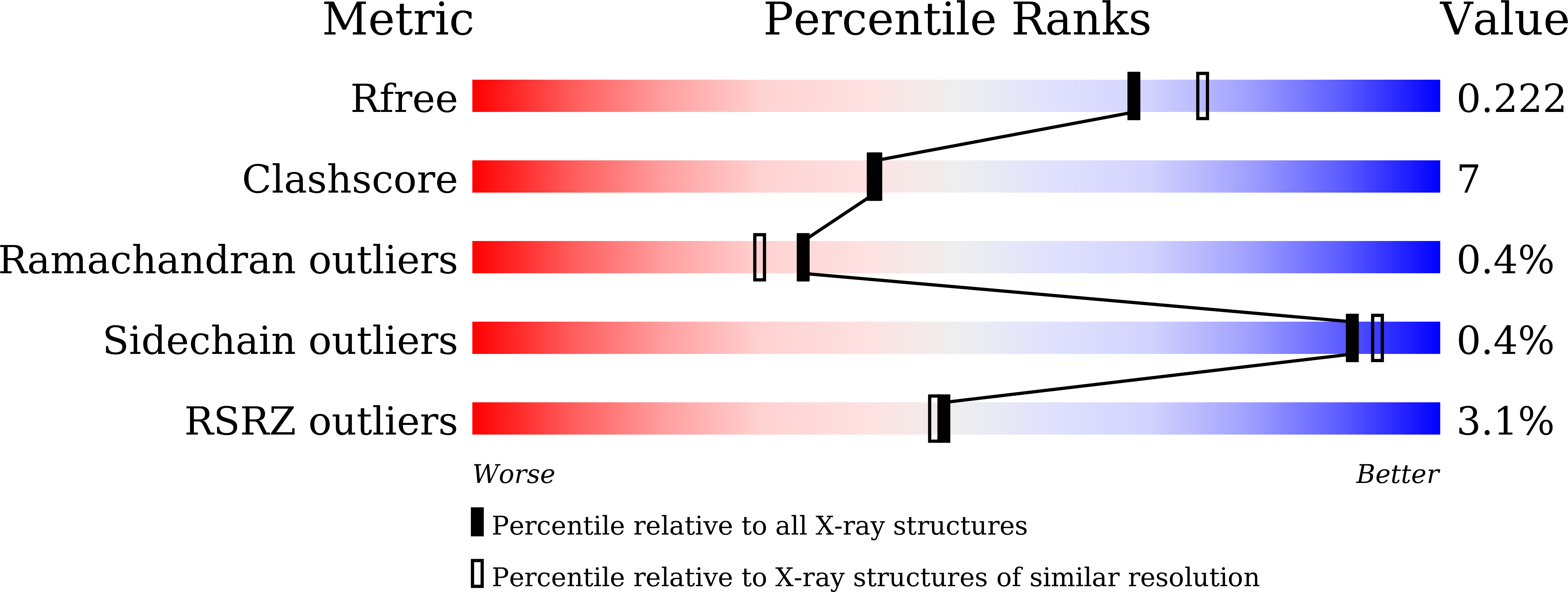
5Y8E, 5Y8F - PubMed Abstract:
SEFIR domain-containing proteins are crucial for mammalian adaptive immunity. As a unique intracellular signaling domain, the SEFIR-SEFIR interactions mediate physical protein-protein interactions in the immune signaling network, especially the IL-17- and IL-25-mediated pathways. However, due to the lack of structural information, the detailed molecular mechanism for SEFIR-SEFIR assembly remains unclear. In the present study, we solved the crystal structures of a prokaryotic SEFIR domain from Bacillus cereus F65185 (BcSEFIR), where the SEFIR domain is located at the N terminus. The structure of BcSEFIR revealed two radically distinct SEFIR-SEFIR interaction modes. In the asymmetric form, the C-terminal tail of one SEFIR binds to the helix αA and βB-αB' segment of the other one, while in the symmetric form, the helices ηC and αE and the DE-segment compose the inter-protomer interface. The C-terminal tail of BcSEFIR, critical for asymmetric interaction, is highly conserved among the SEFIR domains of Act1 orthologs from different species, in particular three absolutely conserved residues that constitute an EXXXXPP motif. In the symmetric interaction mode, the most significant contacts made by residues on helix αE are highly conserved in Act1 SEFIR domains, constituted an RLI/LXE motif. The two novel SEFIR-SEFIR interaction modes might explain the structural basis for SEFIR domain-mediated complex assembly in signaling pathways.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China; Department of Immunology, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA.