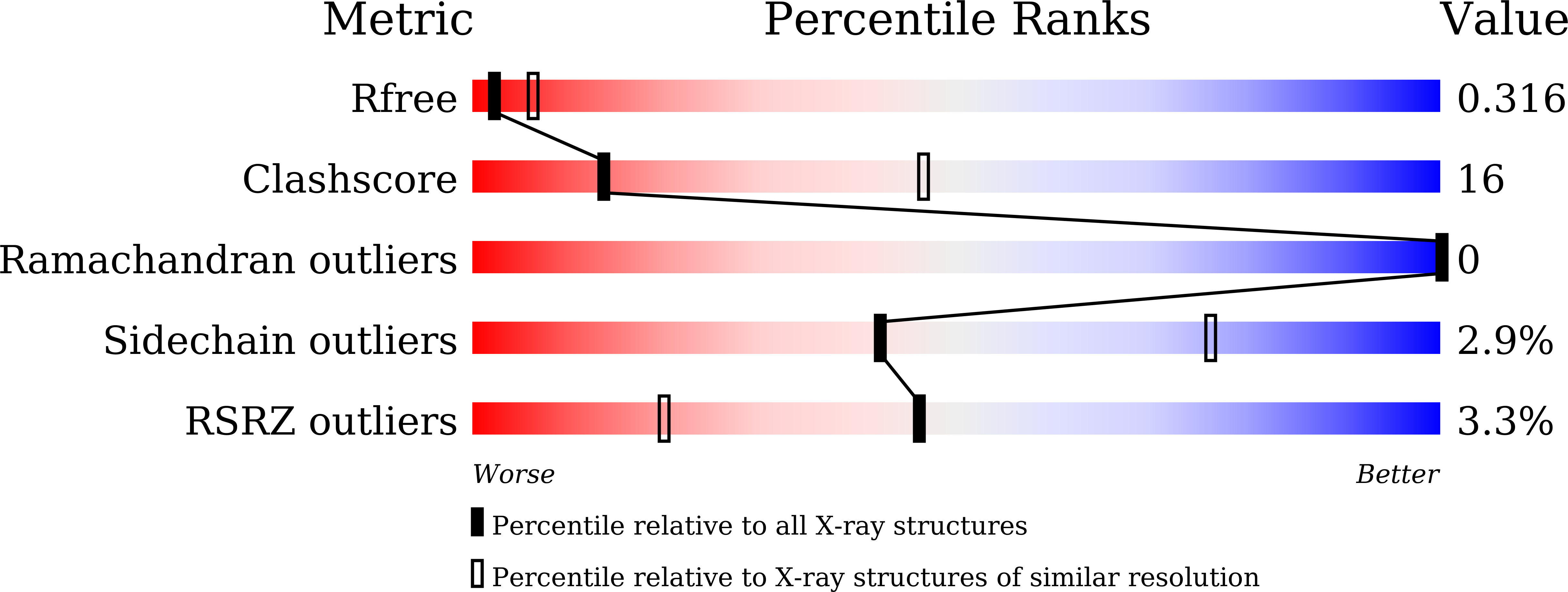
Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Li, Y., Zeng, W., Li, Y., Fan, W., Ma, H., Fan, X., Jiang, J., Brefo-Mensah, E., Zhang, Y., Yang, M., Dong, Z., Palmer, M., Jin, T.(2019) Acta Crystallogr D Struct Biol 75: 772-781
- PubMed: 31373576
- DOI: https://doi.org/10.1107/S205979831901057X
- Primary Citation of Related Structures:
5Y2G - PubMed Abstract:
CAMP factor is a unique α-helical bacterial toxin that is known for its co-hemolytic activity in combination with staphylococcal sphingomyelinase. It was first discovered in the human pathogen Streptococcus agalactiae (also known as group B streptococcus), but homologous genes have been found in many other Gram-positive pathogens. In this study, the efforts that led to the determination of the first structure of a CAMP-family toxin are reported. Initially, it was possible to produce crystals of the native protein which diffracted to near 2.45 Å resolution. However, a series of technical obstacles were encountered on the way to structure determination. Over a period of more than five years, many methods, including selenomethionine labeling, mutations, crystallization chaperones and heavy-atom soaking, were attempted, but these attempts resulted in limited progress. The structure was finally solved using a combination of iodine soaking and molecular replacement using the crystallization chaperone maltose-binding protein (MBP) as a search model. Analysis of native and MBP-tagged CAMP-factor structures identified a conserved interaction interface in the C-terminal domain (CTD). The positively charged surface may be critical for binding to acidic ligands. Furthermore, mutations on the interaction interface at the CTD completely abolished its co-hemolytic activities. This study provides novel insights into the mechanism of the membrane-permeabilizing activity of CAMP factor.
Organizational Affiliation:
Division of Molecular Medicine, Hefei National Laboratory for Physical Sciences at Microscale, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Life Sciences, University of Science and Technology of China, Hefei 230027, People's Republic of China.