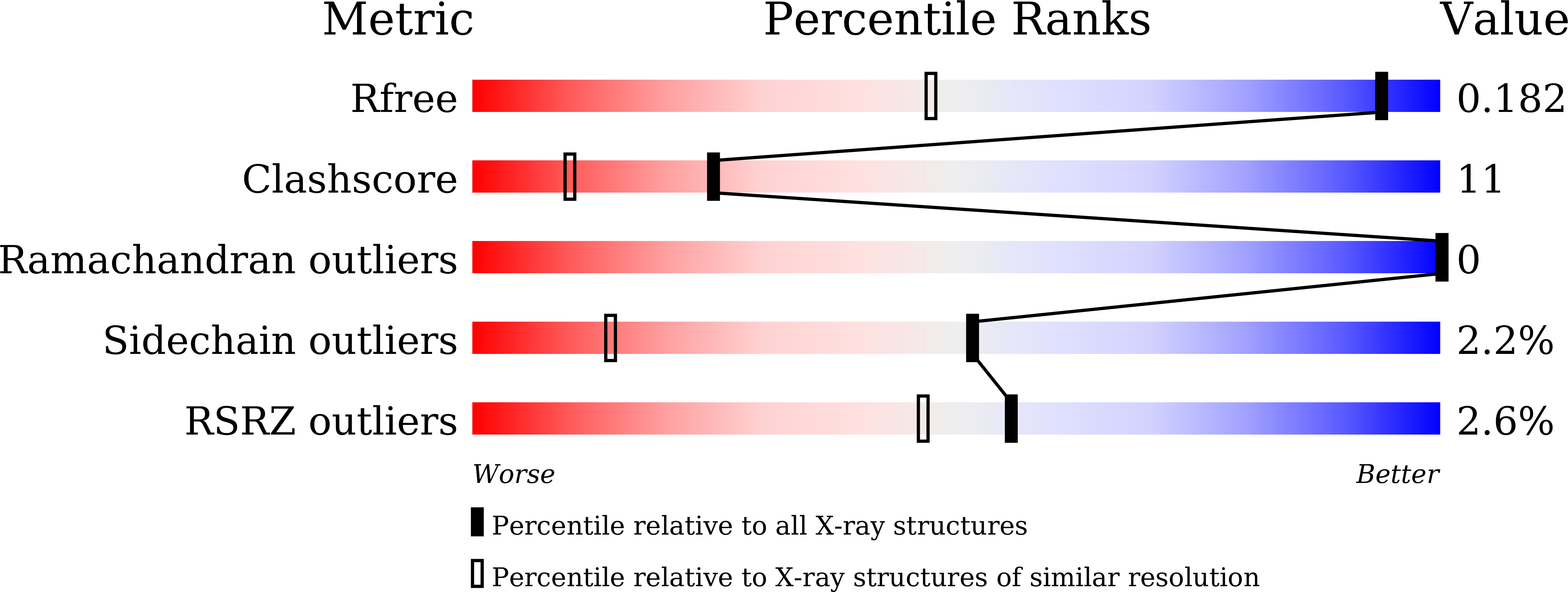
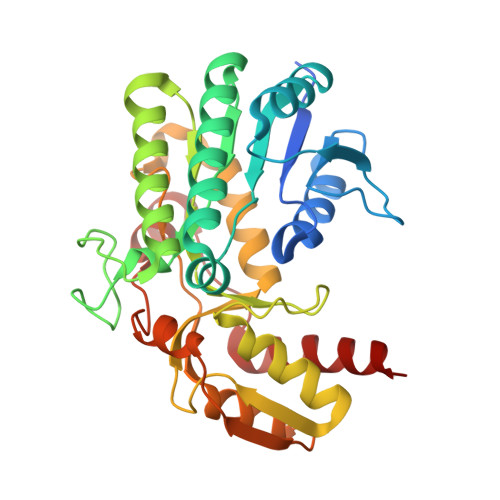
Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Motoyama, T., Nakano, S., Yamamoto, Y., Tokiwa, H., Asano, Y., Ito, S.(2017) Biochemistry 56: 5758-5770
- PubMed: 28992410
- DOI: https://doi.org/10.1021/acs.biochem.7b00832
- Primary Citation of Related Structures:
5Y1D, 5Y1E, 5Y1F, 5Y1G - PubMed Abstract:
A short chain dehydrogenase like l-threonine 3-dehydrogenase (SDR-TDH) from metagenome data (mtTDH) was identified by database mining. Its enzymatic properties suggested that mtTDH has unique characteristics relative to other SDR-TDHs, including two mesophilic and thermophilic SDR-TDHs identified in this study. The activation energy of mtTDH was the lowest (29.6 kJ/mol) of those of the SDR-TDHs, indicating that it is a psychrophilic enzyme. Size-exclusion chromatography analysis revealed mtTDH is a monomer. Crystal structures of mtTDH in apo, binary, and two ternary complexes (l-Ser- and l-Thr-soaked forms) were determined at resolutions of 1.25-1.9 Å. Structural and computational analysis revealed the molecular mechanism of switching between the open and closed states induced by substrate binding and product release. Furthermore, six residues and two water molecules at the active site contributing to product release were assigned. The residues could be categorized into two groups on the basis of the enzymatic properties of their variants: S111, Y136, and T177 and S74, T178, and D179. The former group appeared to affect l-Thr dehydrogenation directly, because the k cat value of their variants was >80-fold lower than that of wild-type mtTDH. On the other hand, the latter group contributes to switching between the open and closed states, which is important for the high substrate specificity of SDR-TDH for l-Thr: the k cat and K m toward l-Thr values of variants in these residues could not be determined because the initial velocity was unsaturated at high concentrations of l-Thr. On the basis of these findings, we proposed a product release mechanism for SDR-TDH associated with specific structural changes.
Organizational Affiliation:
Graduate Division of Nutritional and Environmental Sciences, University of Shizuoka , 52-1 Yada, Suruga-ku, Shizuoka 422-8526, Japan.