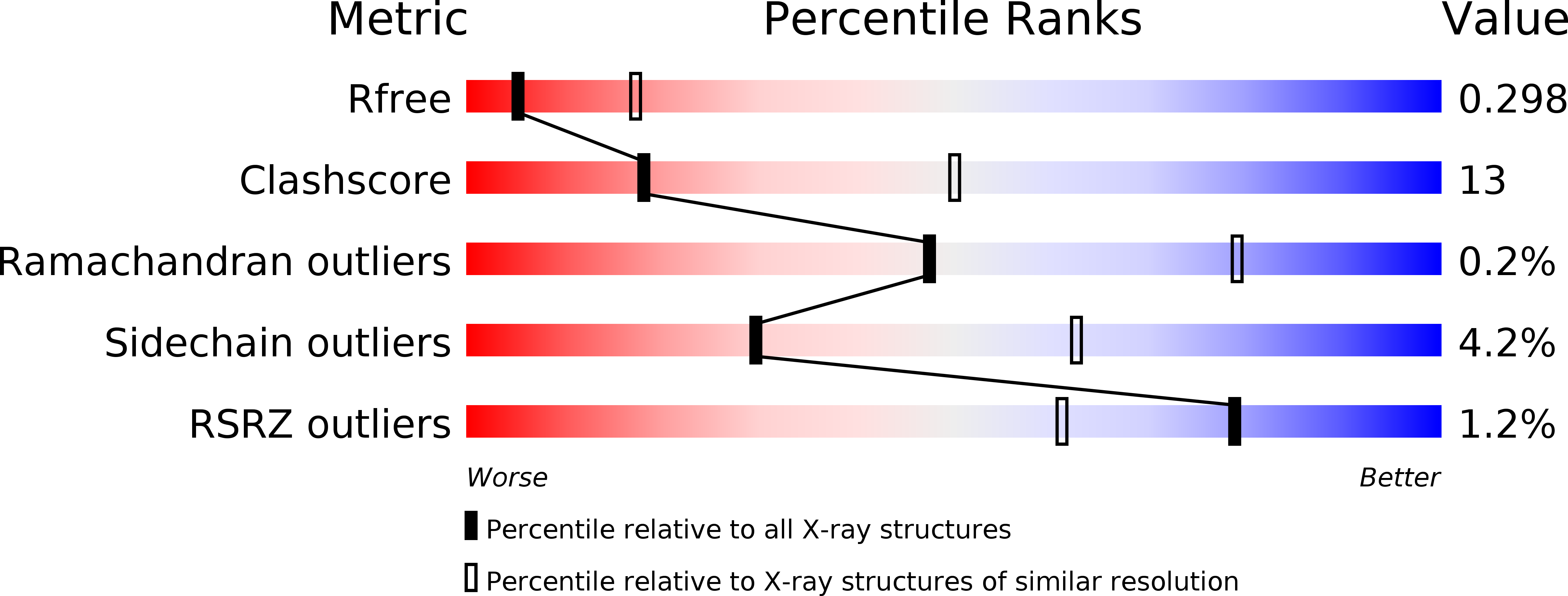
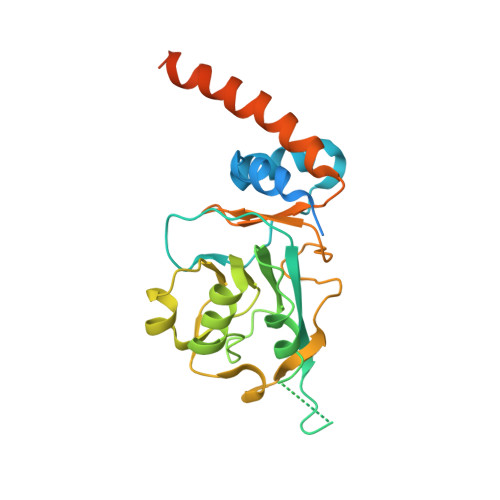
Crystal structure of Streptomyces coelicolor RraAS2, an unusual member of the RNase E inhibitor RraA protein family
Park, N., Heo, J., Song, S., Jo, I., Lee, K., Ha, N.C.(2017) J Microbiol 55: 388-395
- PubMed: 28455590
- DOI: https://doi.org/10.1007/s12275-017-7053-8
- Primary Citation of Related Structures:
5X15 - PubMed Abstract:
Bacterial ribonuclease E (RNase E) plays a crucial role in the processing and decay of RNAs. A small protein named RraA negatively regulates the activity of RNase E via protein-protein interaction in various bacteria. Recently, RraAS1 and RraAS2, which are functional homologs of RraA from Escherichia coli, were identified in the Gram-positive species Streptomyces coelicolor. RraAS1 and RraAS2 inhibit RNase ES ribonuclease activity in S. coelicolor. RraAS1 and RraAS2 have a C-terminal extension region unlike typical bacterial RraA proteins. In this study, we present the crystal structure of RraAS2, exhibiting a hexamer arranged in a dimer of trimers, consistent with size exclusion chromatographic results. Importantly, the C-terminal extension region formed a long α-helix at the junction of the neighboring subunit, which is similar to the trimeric RraA orthologs from Saccharomyces cerevisiae. Truncation of the C-terminal extension region resulted in loss of RNase ES inhibition, demonstrating its crucial role. Our findings present the first bacterial RraA that has a hexameric assembly with a C-terminal extension α-helical region, which plays an essential role in the regulation of RNase ES activity in S. coelicolor.
Organizational Affiliation:
Department of Agricultural Biotechnology, Center for Food Safety and Toxicology, Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul, 08826, Republic of Korea.