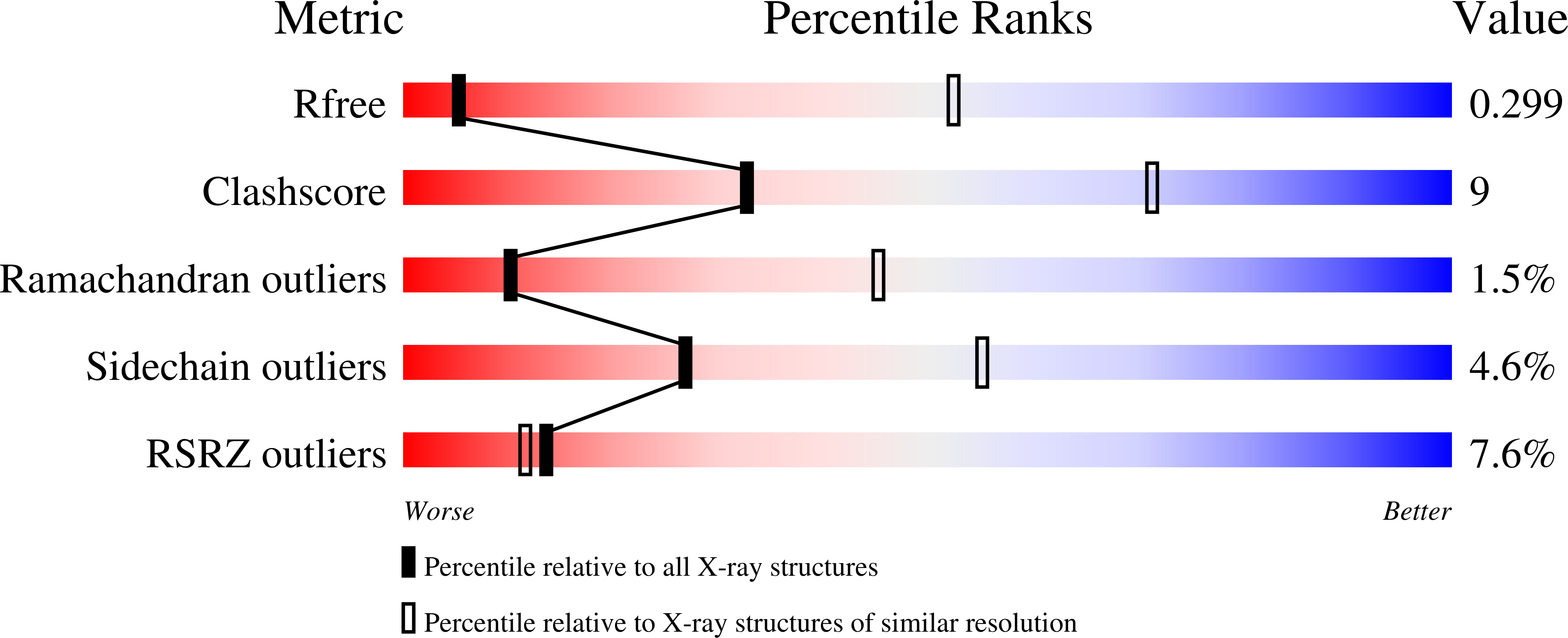
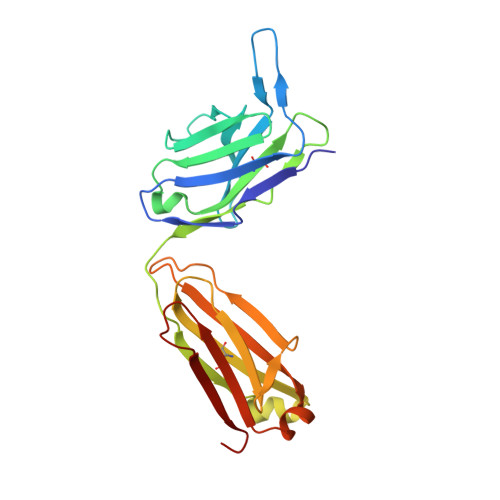
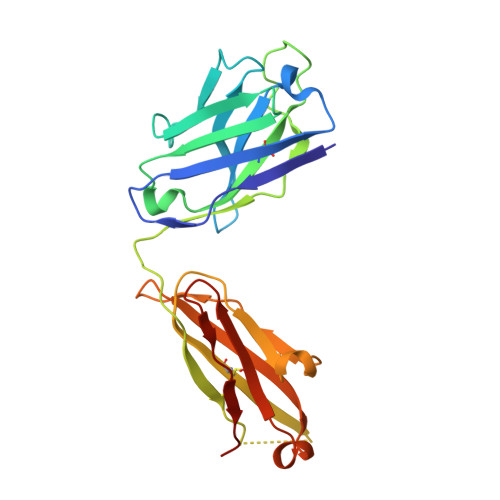
Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Clark, L.E., Mahmutovic, S., Raymond, D.D., Dilanyan, T., Koma, T., Manning, J.T., Shankar, S., Levis, S.C., Briggiler, A.M., Enria, D.A., Wucherpfennig, K.W., Paessler, S., Abraham, J.(2018) Nat Commun 9: 1884-1884
- PubMed: 29760382
- DOI: https://doi.org/10.1038/s41467-018-04271-z
- Primary Citation of Related Structures:
5W1G, 5W1K, 5W1M - PubMed Abstract:
While five arenaviruses cause human hemorrhagic fevers in the Western Hemisphere, only Junin virus (JUNV) has a vaccine. The GP1 subunit of their envelope glycoprotein binds transferrin receptor 1 (TfR1) using a surface that substantially varies in sequence among the viruses. As such, receptor-mimicking antibodies described to date are type-specific and lack the usual breadth associated with this mode of neutralization. Here we isolate, from the blood of a recipient of the live attenuated JUNV vaccine, two antibodies that cross-neutralize Machupo virus with varying efficiency. Structures of GP1-Fab complexes explain the basis for efficient cross-neutralization, which involves avoiding receptor mimicry and targeting a conserved epitope within the receptor-binding site (RBS). The viral RBS, despite its extensive sequence diversity, is therefore a target for cross-reactive antibodies with activity against New World arenaviruses of public health concern.
Organizational Affiliation:
Department of Microbiology and Immunobiology, Harvard Medical School, Boston, MA, 02115, USA.