Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
Iwatani-Yoshihara, M., Ito, M., Klein, M.G., Yamamoto, T., Yonemori, K., Tanaka, T., Miwa, M., Morishita, D., Endo, S., Tjhen, R., Qin, L., Nakanishi, A., Maezaki, H., Kawamoto, T.(2017) J Med Chem 60: 5759-5771
- PubMed: 28586220
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00461
- Primary Citation of Related Structures:
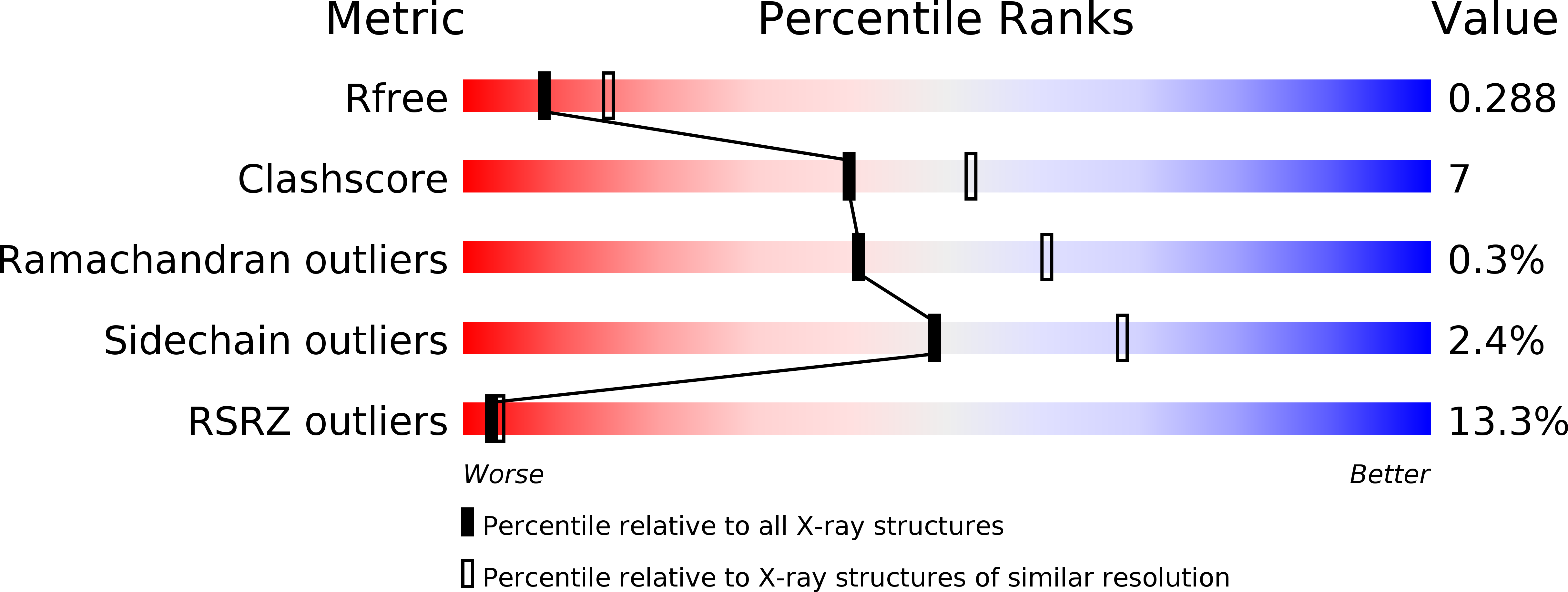
5URJ, 5URK, 5URM - PubMed Abstract:
Brr2 is an RNA helicase belonging to the Ski2-like subfamily and an essential component of spliceosome. Brr2 catalyzes an ATP-dependent unwinding of the U4/U6 RNA duplex, which is a critical step for spliceosomal activation. An HTS campaign using an RNA-dependent ATPase assay and initial SAR study identified two different Brr2 inhibitors, 3 and 12. Cocrystal structures revealed 3 binds to an unexpected allosteric site between the C-terminal and the N-terminal helicase cassettes, while 12 binds an RNA-binding site inside the N-terminal cassette. Selectivity profiling indicated the allosteric inhibitor 3 is more Brr2-selective than the RNA site binder 12. Chemical optimization of 3 using SBDD culminated in the discovery of the potent and selective Brr2 inhibitor 9 with helicase inhibitory activity. Our findings demonstrate an effective strategy to explore selective inhibitors for helicases, and 9 could be a promising starting point for exploring molecular probes to elucidate biological functions and the therapeutic relevance of Brr2.
Organizational Affiliation:
Department of Structural Biology, Takeda California Inc. , 10410 Science Center Drive, San Diego, California 92121, United States.