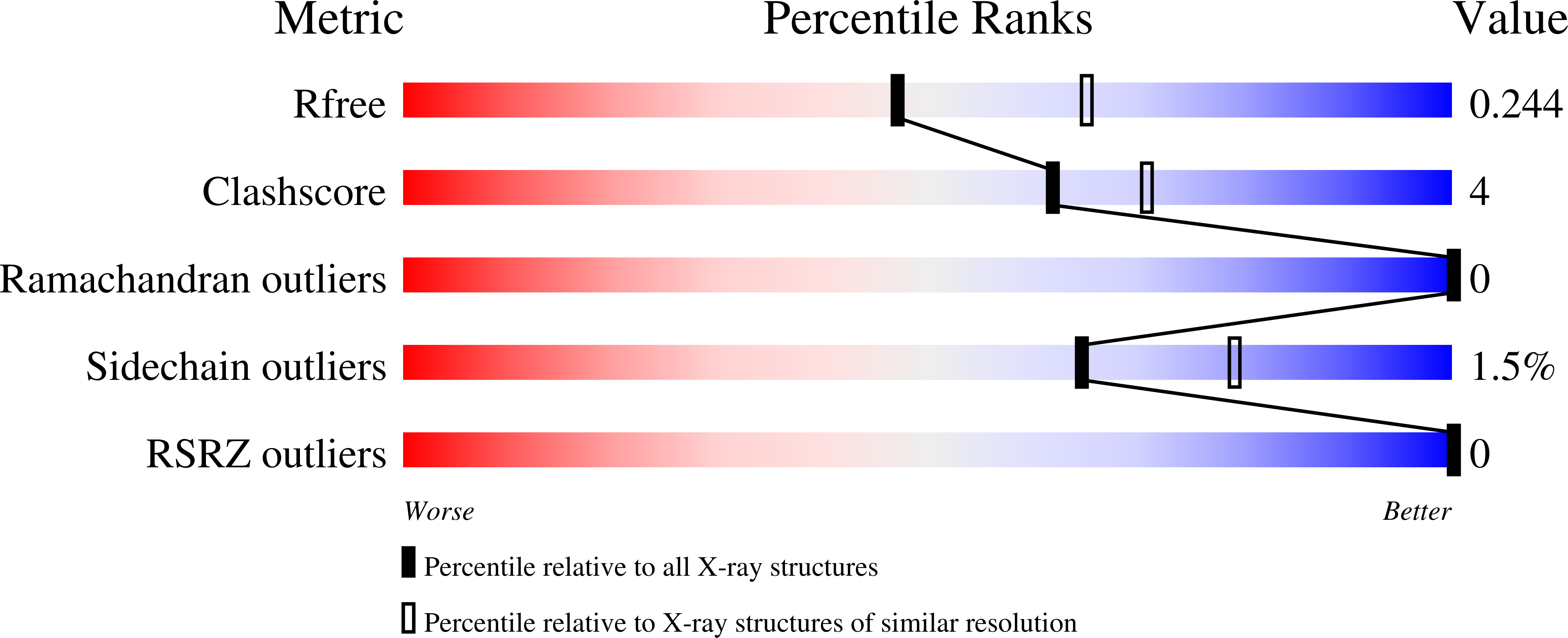
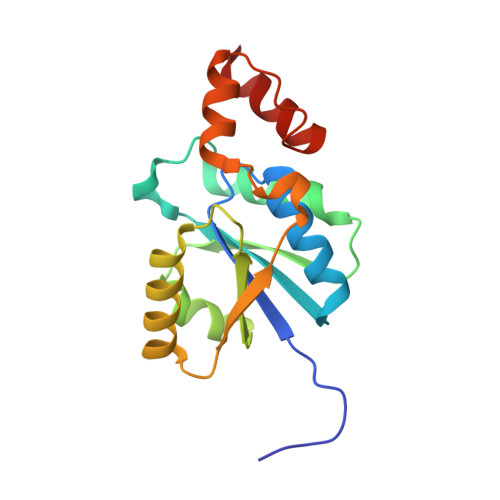
Crystal structure of a phosphopantetheine adenylyltransferase (CoaD, PPAT) from Pseudomonas aeruginosa bound to dephospho coenzyme A
Edwards, T.E., Davies, D.R., Fairman, J.W., Lorimer, D., Seattle Structural Genomics Center for Infectious Disease (SSGCID)To be published.