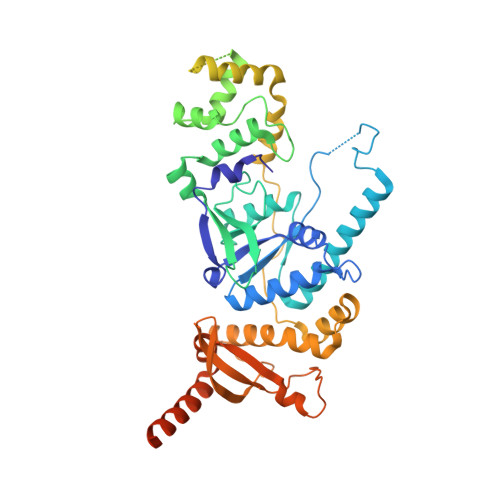
Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage.
Lee, S.H., Princz, L.N., Klugel, M.F., Habermann, B., Pfander, B., Biertumpfel, C.(2015) Elife 4
- PubMed: 26682650
- DOI: https://doi.org/10.7554/eLife.12256
- Primary Citation of Related Structures:
5T9J - PubMed Abstract:
Holliday junctions (HJs) are key DNA intermediates in homologous recombination. They link homologous DNA strands and have to be faithfully removed for proper DNA segregation and genome integrity. Here, we present the crystal structure of human HJ resolvase GEN1 complexed with DNA at 3.0 Å resolution. The GEN1 core is similar to other Rad2/XPG nucleases. However, unlike other members of the superfamily, GEN1 contains a chromodomain as an additional DNA interaction site. Chromodomains are known for their chromatin-targeting function in chromatin remodelers and histone(de)acetylases but they have not previously been found in nucleases. The GEN1 chromodomain directly contacts DNA and its truncation severely hampers GEN1's catalytic activity. Structure-guided mutations in vitro and in vivo in yeast validated our mechanistic findings. Our study provides the missing structure in the Rad2/XPG family and insights how a well-conserved nuclease core acquires versatility in recognizing diverse substrates for DNA repair and maintenance.
Organizational Affiliation:
Department of Structural Cell Biology, Molecular Mechanisms of DNA Repair, Max Planck Institute of Biochemistry, Martinsried, Germany.