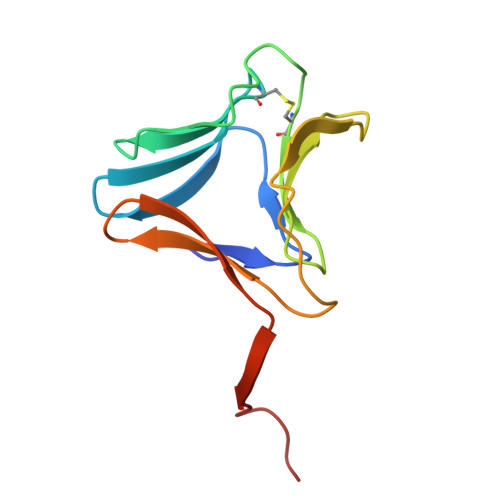
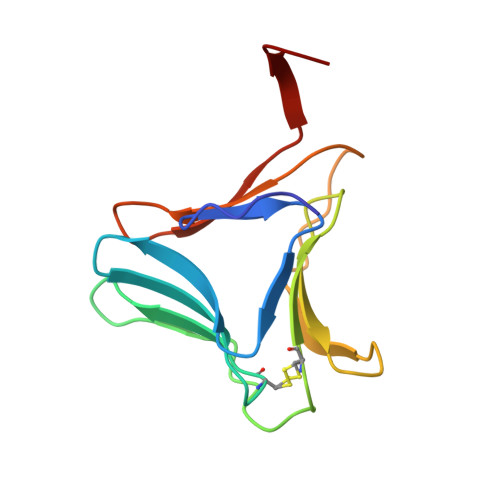
High-resolution crystal structures of Colocasia esculenta tarin lectin.
Pereira, P.R., Meagher, J.L., Winter, H.C., Goldstein, I.J., Paschoalin, V.M., Silva, J.T., Stuckey, J.A.(2017) Glycobiology 27: 50-56
- PubMed: 27558840
- DOI: https://doi.org/10.1093/glycob/cww083
- Primary Citation of Related Structures:
5T1X, 5T20 - PubMed Abstract:
Tarin, the Colocasia esculenta lectin from the superfamily of α-d-mannose-specific plant bulb lectins, is a tetramer of 47 kDa composed of two heterodimers. Each heterodimer possesses homologous monomers of ~11.9 (A chain) and ~12.7 (B chain) kDa. The structures of apo and carbohydrate-bound tarin were solved to 1.7 Å and 1.91 Å, respectively. Each tarin monomer forms a canonical β-prism II fold, common to all members of Galanthus nivalis agglutinin (GNA) family, which is partially stabilized by a disulfide bond and a conserved hydrophobic core. The heterodimer is formed through domain swapping involving the C-terminal β-strand and the β-sheet on face I of the prism. The tetramer is assembled through the dimerization of the B chains from heterodimers involving face II of each prism. The 1.91 Å crystal structure of tarin bound to Manα(1,3)Manα(1,6)Man reveals an expanded carbohydrate-binding sequence (QxDxNxVxYx 4/6 WX) on face III of the β-prism. Both monomers possess a similar fold, except for the length of the loop, which begins after the conserved tyrosine and creates the binding pocket for the α(1,6)-terminal mannose. This loop differs in size and amino-acid composition from 10 other β-prism II domain proteins, and may confer carbohydrate-binding specificity among members of the GNA-related lectin family.
Organizational Affiliation:
Centro de Tecnologia, Universidade Federal do Rio de Janeiro (UFRJ), Avenida Athos da Silveira Ramos, 149., Rio de Janeiro 21941-909, Brazil.