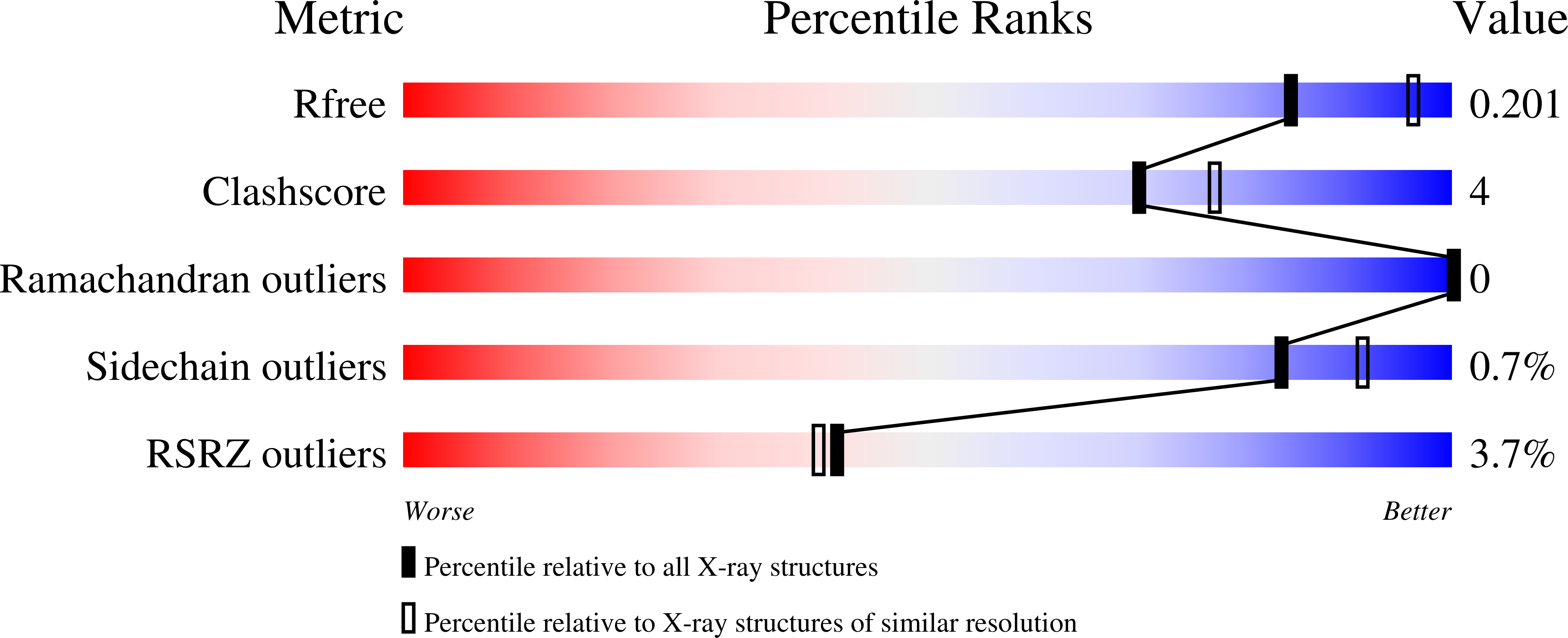
Structure of the Cyanuric Acid Hydrolase TrzD Reveals Product Exit Channel.
Bera, A.K., Aukema, K.G., Elias, M., Wackett, L.P.(2017) Sci Rep 7: 45277-45277
- PubMed: 28345631
- DOI: https://doi.org/10.1038/srep45277
- Primary Citation of Related Structures:
5T13 - PubMed Abstract:
Cyanuric acid hydrolases are of industrial importance because of their use in aquatic recreational facilities to remove cyanuric acid, a stabilizer for the chlorine. Degradation of excess cyanuric acid is necessary to maintain chlorine disinfection in the waters. Cyanuric acid hydrolase opens the cyanuric acid ring hydrolytically and subsequent decarboxylation produces carbon dioxide and biuret. In the present study, we report the X-ray structure of TrzD, a cyanuric acid hydrolase from Acidovorax citrulli. The crystal structure at 2.19 Å resolution shows a large displacement of the catalytic lysine (Lys163) in domain 2 away from the active site core, whereas the two other active site lysines from the two other domains are not able to move. The lysine displacement is proposed here to open up a channel for product release. Consistent with that, the structure also showed two molecules of the co-product, carbon dioxide, one in the active site and another trapped in the proposed exit channel. Previous data indicated that the domain 2 lysine residue plays a role in activating an adjacent serine residue carrying out nucleophilic attack, opening the cyanuric acid ring, and the mobile lysine guides products through the exit channel.
Organizational Affiliation:
BioTechnology Institute, University of Minnesota, St. Paul, MN 55108, USA.