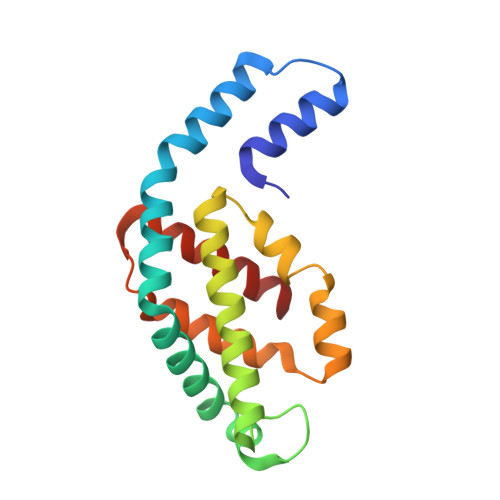
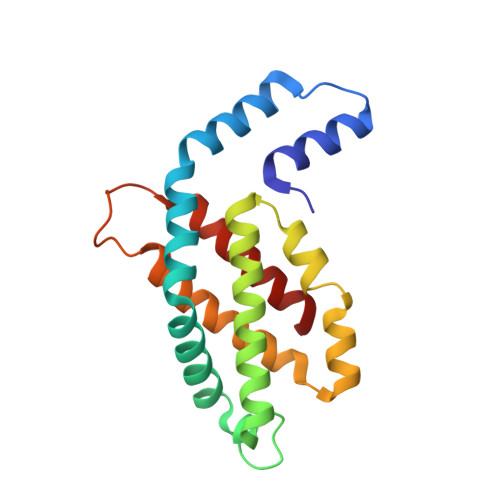
Structural heterogeneity leads to functional homogeneity in A. marina phycocyanin.
Bar-Zvi, S., Lahav, A., Harris, D., Niedzwiedzki, D.M., Blankenship, R.E., Adir, N.(2018) Biochim Biophys Acta 1859: 544-553
- PubMed: 29704497
- DOI: https://doi.org/10.1016/j.bbabio.2018.04.007
- Primary Citation of Related Structures:
5OOK - PubMed Abstract:
The major light harvesting antenna in all cyanobacterial species is the phycobilisome (PBS). The smallest PBS identified to date is that of Acaryochloris marina (A. marina), composed of a single four-hexamer rod. We have determined the crystal structure of phycocyanin (AmPC), the major component of the A. marina PBS (AmPBS) to 2.1 Å. The basic unit of the AmPC is a heterodimer of two related subunits (α and β), and we show that the asymmetric unit contains a superposition of two α and two β isoforms, the products of the simultaneous expression of different genes. This is the first time to our knowledge that isolated proteins crystallized with such identifiable heterogeneity. We believe that the presence of the different isoforms allows the AmPBS to have a significant bathochromic shift in its fluorescence emission spectrum, allowing, in the total absence of allophycocyanin, a better overlap with absorption of the chlorophyll d-containing reaction centers. We show that this bathochromic shift exists in intact AmPBS as well as in its disassembled components, thus suggesting that AmPC can efficiently serve as the AmPBS terminal emitter.
Organizational Affiliation:
Schulich Faculty of Chemistry, Technion-Israel Institute of Technology, Haifa 32000, Israel.