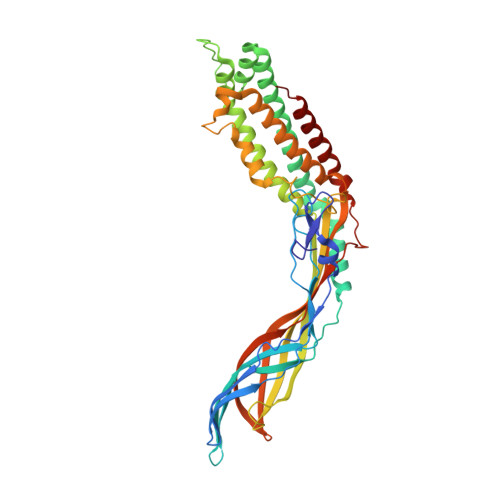
Crystal Structure of the Maturation Protein from Bacteriophage Q beta.
Rumnieks, J., Tars, K.(2017) J Mol Biol 429: 688-696
- PubMed: 28111107
- DOI: https://doi.org/10.1016/j.jmb.2017.01.012
- Primary Citation of Related Structures:
5MNT - PubMed Abstract:
Virions of the single-stranded RNA bacteriophages contain a single copy of the maturation protein, which is bound to the phage genome and is required for the infectivity of the particles. The maturation protein mediates the adsorption of the virion to bacterial pili and the subsequent release and penetration of the genome into the host cell. Here, we report a crystal structure of the maturation protein from bacteriophage Qβ. The protein has a bent, highly asymmetric shape and spans 110Å in length. Apart from small local substructures, the overall fold of the maturation protein does not resemble that of other known proteins. The protein is organized in two distinct regions, an α-helical part with a four-helix core, and a β stranded part that contains a seven-stranded sheet in the central part and a five-stranded sheet at the tip of the protein. The Qβ maturation protein has two distinct, positively charged areas at opposite sides of the α-helical part, which are involved in genomic RNA binding. The maturation protein binds to each of the surrounding coat protein dimers in the capsid differently, and the interaction is considerably weaker compared to coat protein interdimer contacts. The coat protein- or RNA-binding residues are not preserved among different ssRNA phage maturation proteins; instead, the distal end of the α-helical part is the most evolutionarily conserved, suggesting the importance of this region for maintaining the functionality of the protein.
Organizational Affiliation:
Biomedical Research and Study Center, Ratsupites 1, LV1067 Riga, Latvia.