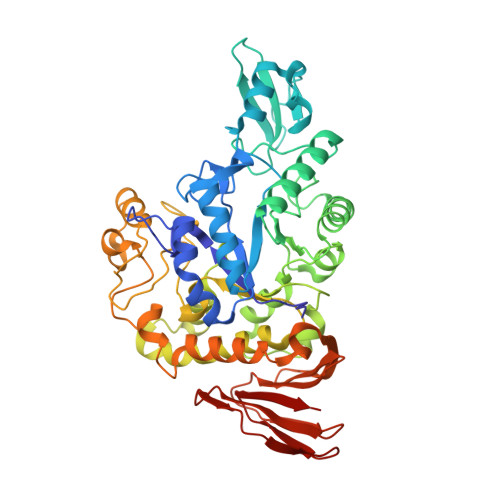
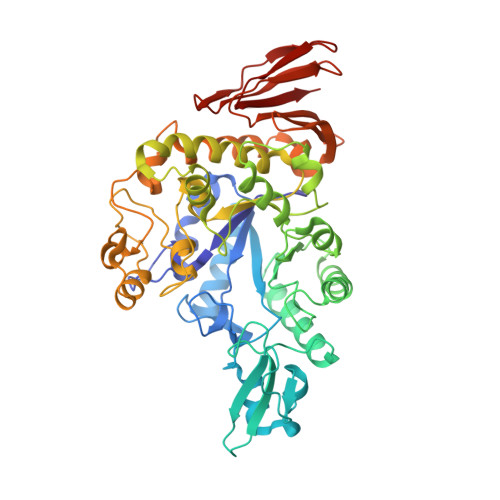
Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Hameed, U., Price, I., Ke, A., Wilson, D.B., Mirza, O.(2017) Biochim Biophys Acta 1865: 1237-1245
- PubMed: 28648523
- DOI: https://doi.org/10.1016/j.bbapap.2017.06.015
- Primary Citation of Related Structures:
5M99 - PubMed Abstract:
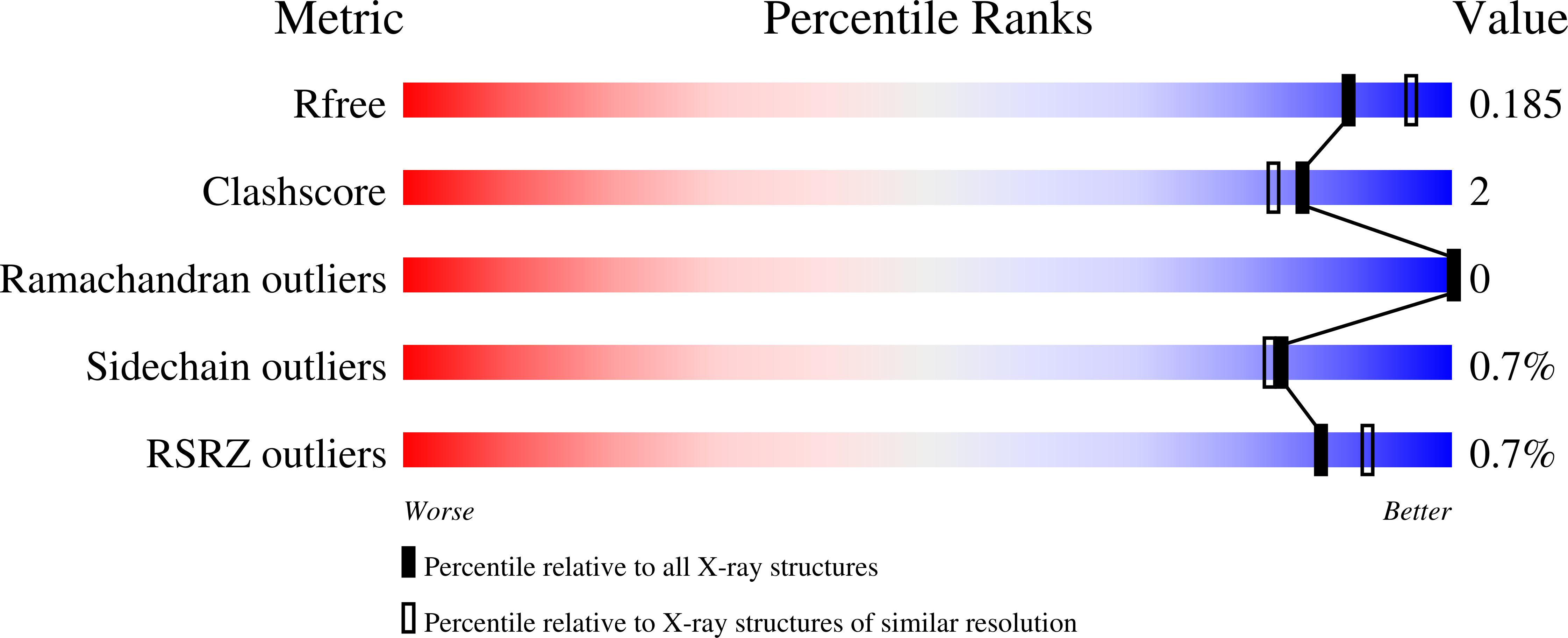
Thermostable α-amylases have many industrial applications and are therefore continuously explored from novel sources. We present the characterization of a novel putative α-amylase gene product (Tp-AmyS) cloned from Thermotoga petrophila. The purified recombinant enzyme is highly thermostable and able to hydrolyze starch into dextrin between 90 and 100°C, with optimum activity at 98°C and pH8.5. The activity increased in the presence of Rb 1+ , K 1+ and Ca 2+ ions, whereas other ions inhibited activity. The crystal structure of Tp-AmyS at 1.7Å resolution showed common features of the GH-13 family, however was apparently found to be a dimer. Several residues from one monomer interacted with a docked acarbose, an inhibitor of Tp-AmyS, in the other monomer, suggesting catalytic cooperativity within the dimer. The most striking feature of the dimer was that it resembled the dimerization of salivary amylase from a previous crystal structure, and thus could be a functional feature of some amylases.
Organizational Affiliation:
Institute of Industrial Biotechnology, Government College University, Lahore, 54000, Pakistan.