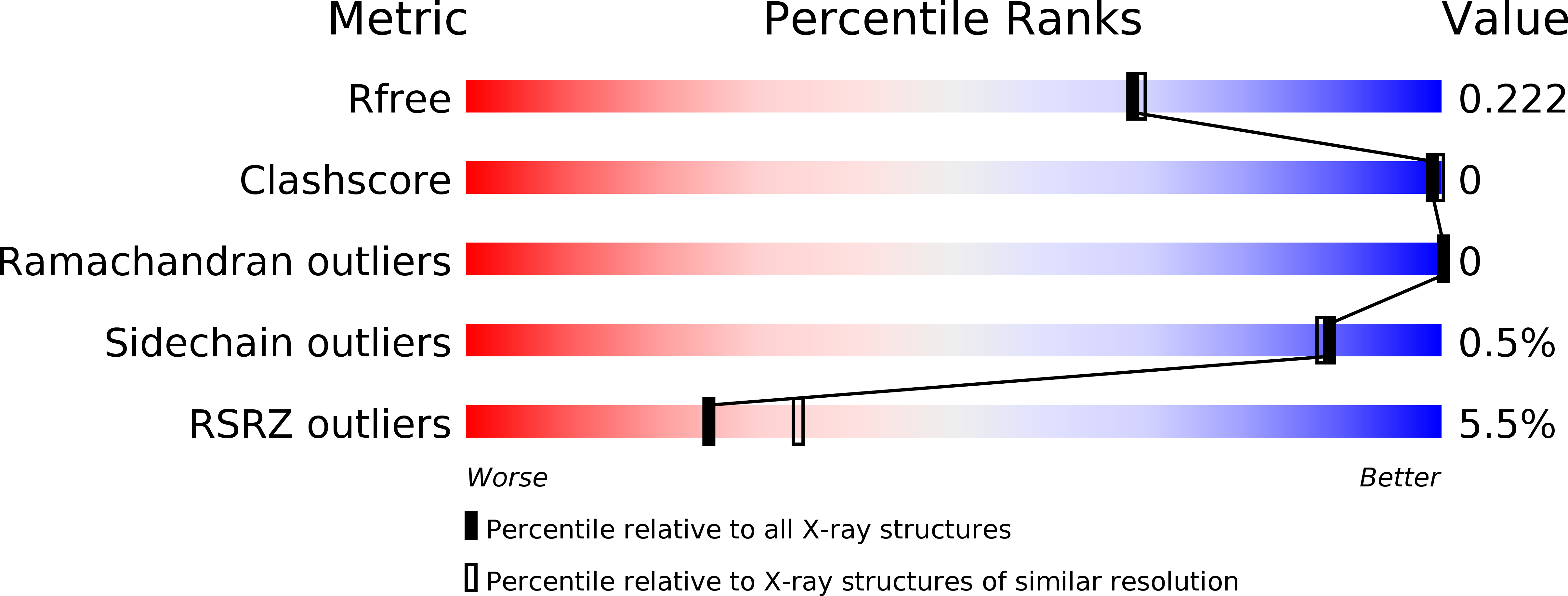
Crystal structures of a yeast 14-3-3 protein from Lachancea thermotolerans in the unliganded form and bound to a human lipid kinase PI4KB-derived peptide reveal high evolutionary conservation.
Eisenreichova, A., Klima, M., Boura, E.(2016) Acta Crystallogr F Struct Biol Commun 72: 799-803
- PubMed: 27827352
- DOI: https://doi.org/10.1107/S2053230X16015053
- Primary Citation of Related Structures:
5LVZ - PubMed Abstract:
14-3-3 proteins bind phosphorylated binding partners to regulate several of their properties, including enzymatic activity, stability and subcellular localization. Here, two crystal structures are presented: the crystal structures of the 14-3-3 protein (also known as Bmh1) from the yeast Lachancea thermotolerans in the unliganded form and bound to a phosphopeptide derived from human PI4KB (phosphatidylinositol 4-kinase B). The structures demonstrate the high evolutionary conservation of ligand recognition by 14-3-3 proteins. The structural analysis suggests that ligand recognition by 14-3-3 proteins evolved very early in the evolution of eukaryotes and remained conserved, underlying the importance of 14-3-3 proteins in physiology.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, v.v.i., Flemingovo nam. 2, 166 10 Prague 6, Czech Republic.