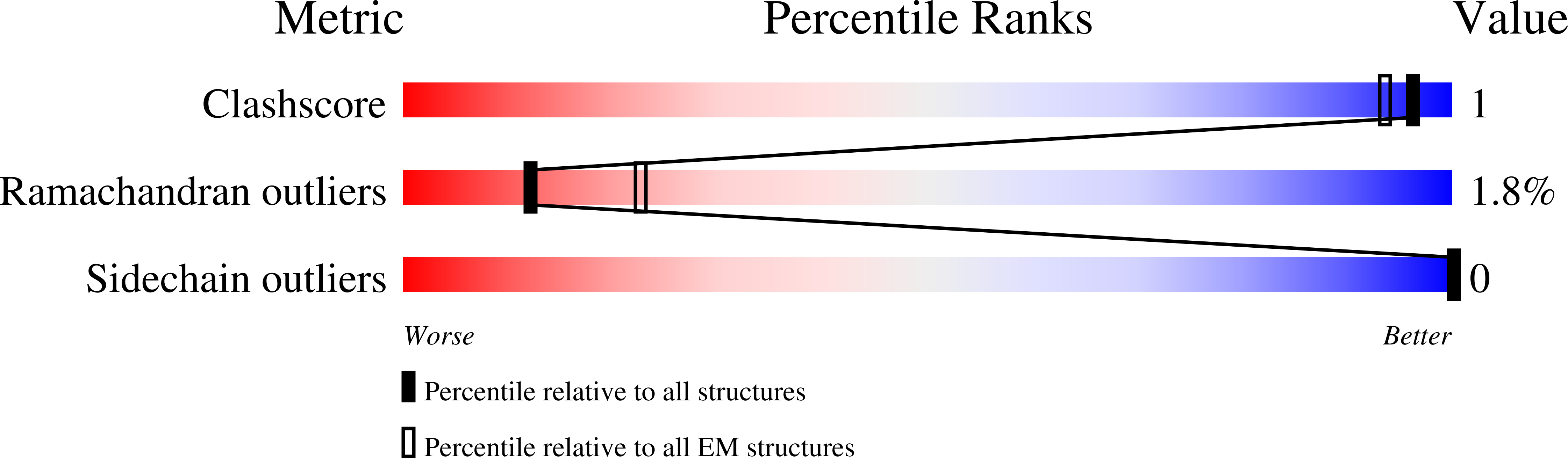Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Vinothkumar, K.R., Montgomery, M.G., Liu, S., Walker, J.E.(2016) Proc Natl Acad Sci U S A 113: 12709-12714
- PubMed: 27791192
- DOI: https://doi.org/10.1073/pnas.1615902113
- Primary Citation of Related Structures:
5LQX, 5LQY, 5LQZ - PubMed Abstract:
The structure of the intact monomeric ATP synthase from the fungus, Pichia angusta , has been solved by electron cryo-microscopy. The structure provides insights into the mechanical coupling of the transmembrane proton motive force across mitochondrial membranes in the synthesis of ATP. This mechanism requires a strong and integral stator, consisting of the catalytic α 3 β 3 -domain, peripheral stalk, and, in the membrane domain, subunit a and associated supernumerary subunits, kept in contact with the rotor turning at speeds up to 350 Hz. The stator's integrity is ensured by robust attachment of both the oligomycin sensitivity conferral protein (OSCP) to the catalytic domain and the membrane domain of subunit b to subunit a. The ATP8 subunit provides an additional brace between the peripheral stalk and subunit a. At the junction between the OSCP and the apparently stiff, elongated α-helical b-subunit and associated d- and h-subunits, an elbow or joint allows the stator to bend to accommodate lateral movements during the activity of the catalytic domain. The stator may also apply lateral force to help keep the static a-subunit and rotating c 10 -ring together. The interface between the c 10 -ring and the a-subunit contains the transmembrane pathway for protons, and their passage across the membrane generates the turning of the rotor. The pathway has two half-channels containing conserved polar residues provided by a bundle of four α-helices inclined at ∼30° to the plane of the membrane, similar to those described in other species. The structure provides more insights into the workings of this amazing machine.
Organizational Affiliation:
The Medical Research Council Laboratory of Molecular Biology, Cambridge Biomedical Campus, Cambridge CB2 0QH, United Kingdom.