Structural Basis of Host Autophagy-related Protein 8 (ATG8) Binding by the Irish Potato Famine Pathogen Effector Protein PexRD54.
Maqbool, A., Hughes, R.K., Dagdas, Y.F., Tregidgo, N., Zess, E., Belhaj, K., Round, A., Bozkurt, T.O., Kamoun, S., Banfield, M.J.(2016) J Biol Chem 291: 20270-20282
- PubMed: 27458016
- DOI: https://doi.org/10.1074/jbc.M116.744995
- Primary Citation of Related Structures:
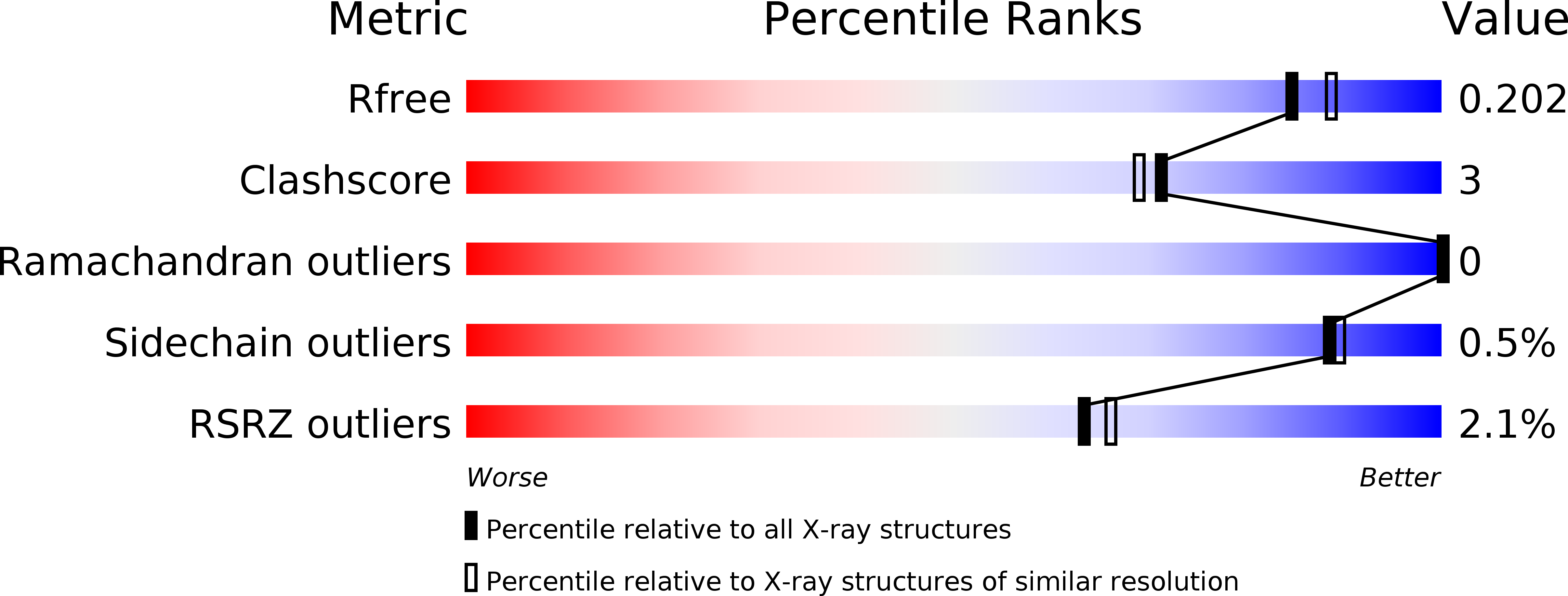
5L7S, 5L83 - PubMed Abstract:
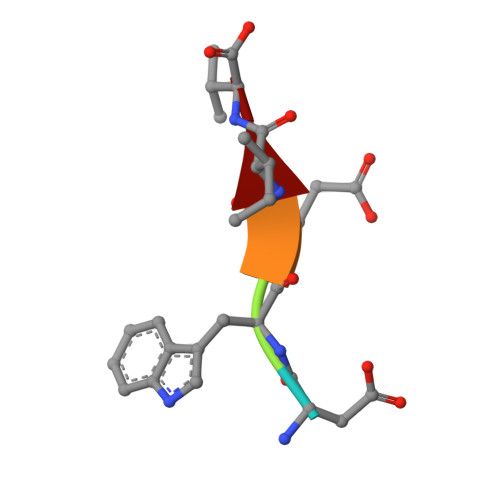
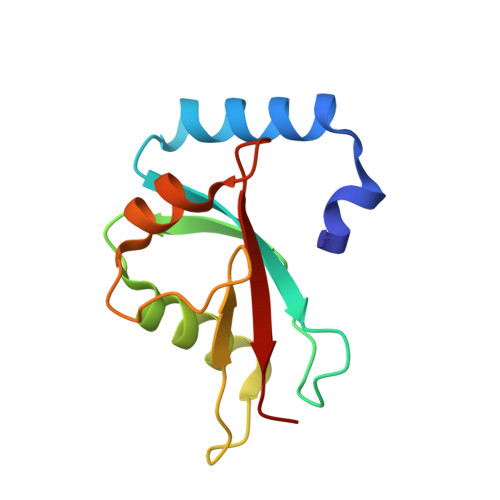
Filamentous plant pathogens deliver effector proteins to host cells to promote infection. The Phytophthora infestans RXLR-type effector PexRD54 binds potato ATG8 via its ATG8 family-interacting motif (AIM) and perturbs host-selective autophagy. However, the structural basis of this interaction remains unknown. Here, we define the crystal structure of PexRD54, which includes a modular architecture, including five tandem repeat domains, with the AIM sequence presented at the disordered C terminus. To determine the interface between PexRD54 and ATG8, we solved the crystal structure of potato ATG8CL in complex with a peptide comprising the effector's AIM sequence, and we established a model of the full-length PexRD54-ATG8CL complex using small angle x-ray scattering. Structure-informed deletion of the PexRD54 tandem domains reveals retention of ATG8CL binding in vitro and in planta This study offers new insights into structure/function relationships of oomycete RXLR effectors and how these proteins engage with host cell targets to promote disease.
Organizational Affiliation:
From the Department of Biological Chemistry, John Innes Centre, and.