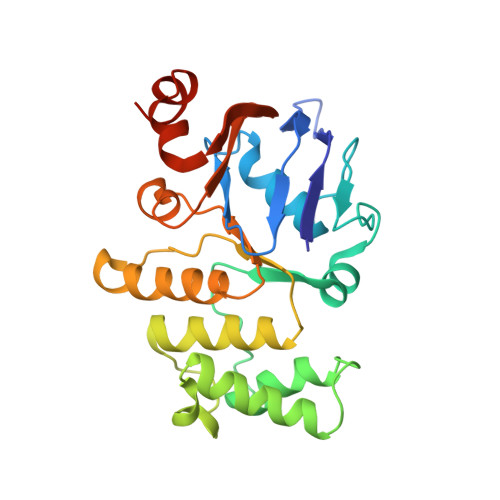
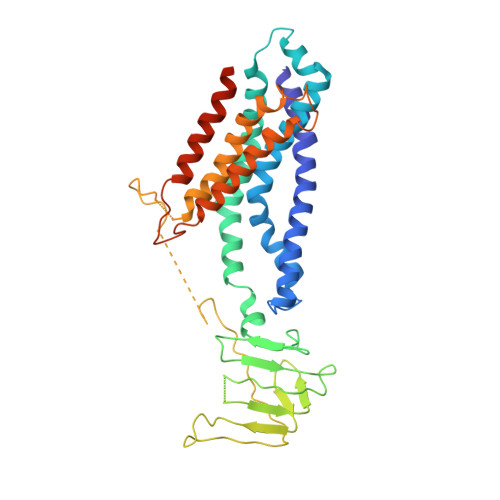
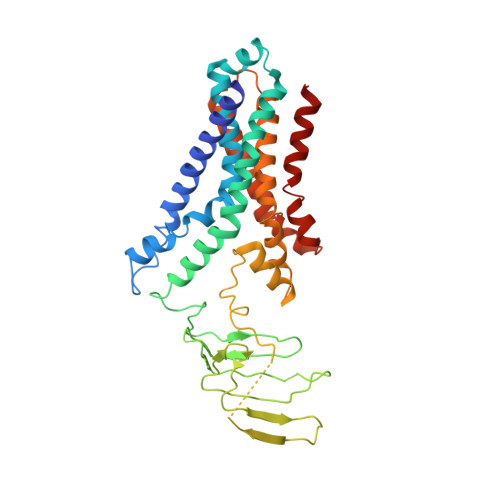
Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG.
Dong, H., Zhang, Z., Tang, X., Paterson, N.G., Dong, C.(2017) Nat Commun 8: 222-222
- PubMed: 28790314
- DOI: https://doi.org/10.1038/s41467-017-00273-5
- Primary Citation of Related Structures:
5L75 - PubMed Abstract:
The cell surface of most Gram-negative bacteria contains lipopolysaccharide that is essential for their viability and drug resistance. A 134-kDa protein complex LptB 2 FG is unique among ATP-binding cassette transporters because it extracts lipopolysaccharide from the external leaflet of the inner membrane and propels it along a filament that extends across the periplasm to directly deliver lipopolysaccharide into the external leaflet of the outer membrane. Here we report the crystal structure of the lipopolysaccharide transporter LptB 2 FG from Klebsiella pneumoniae, in which both LptF and LptG are composed of a β-jellyroll-like periplasmic domain and six α-helical segments in the transmembrane domain. LptF and LptG form a central cavity containing highly conserved hydrophobic residues. Structural and functional studies suggest that LptB 2 FG uses an alternating lateral access mechanism to extract lipopolysaccharide and traffic it along the hydrophobic cavity toward the transporter's periplasmic domains.Lipopolysaccharides (LPS) are synthesized at the periplasmic side of the inner membrane of Gram-negative bacteria and are then extracted by the LptB 2 FG complex during the first step of LPS transport to the outer membrane. Here the authors present the LptB 2 FG structure, which supports an alternating lateral access mechanism for LPS extraction.
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu, 610041, China.