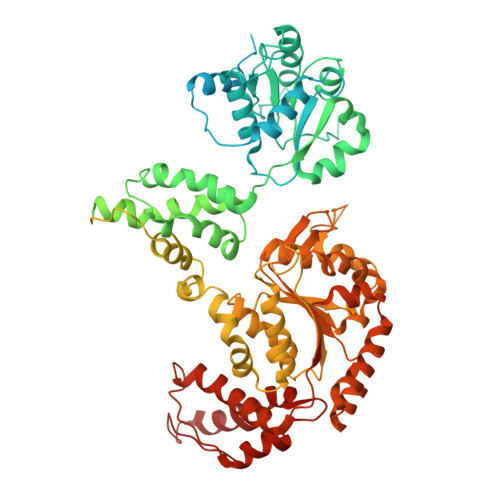
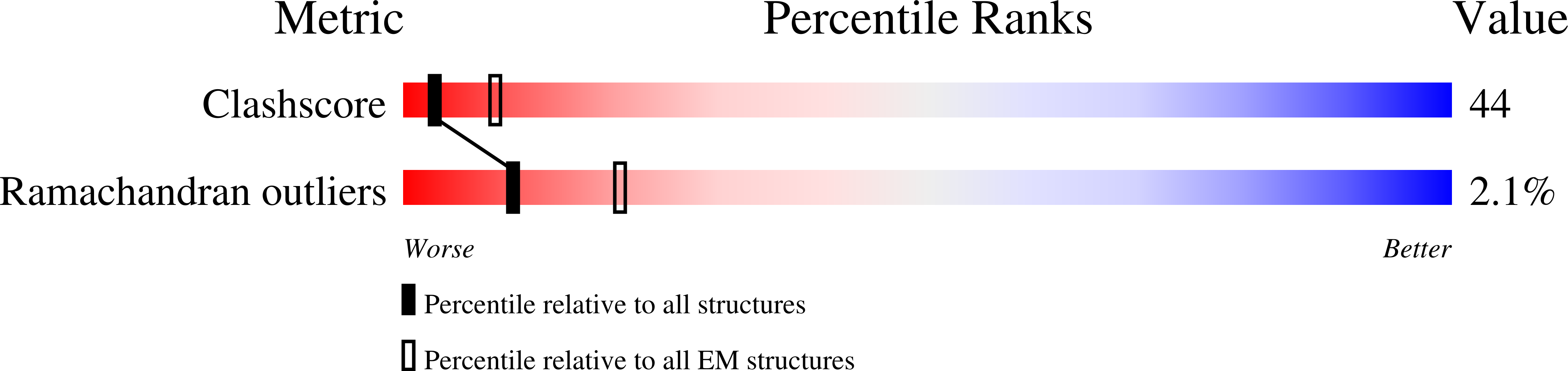Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation.
Yokom, A.L., Gates, S.N., Jackrel, M.E., Mack, K.L., Su, M., Shorter, J., Southworth, D.R.(2016) Nat Struct Mol Biol 23: 830-837
- PubMed: 27478928
- DOI: https://doi.org/10.1038/nsmb.3277
- Primary Citation of Related Structures:
5KNE - PubMed Abstract:
Hsp104, a conserved AAA+ protein disaggregase, promotes survival during cellular stress. Hsp104 remodels amyloids, thereby supporting prion propagation, and disassembles toxic oligomers associated with neurodegenerative diseases. However, a definitive structural mechanism for its disaggregase activity has remained elusive. We determined the cryo-EM structure of wild-type Saccharomyces cerevisiae Hsp104 in the ATP state, revealing a near-helical hexamer architecture that coordinates the mechanical power of the 12 AAA+ domains for disaggregation. An unprecedented heteromeric AAA+ interaction defines an asymmetric seam in an apparent catalytic arrangement that aligns the domains in a two-turn spiral. N-terminal domains form a broad channel entrance for substrate engagement and Hsp70 interaction. Middle-domain helices bridge adjacent protomers across the nucleotide pocket, thus explaining roles in ATP hydrolysis and protein disaggregation. Remarkably, substrate-binding pore loops line the channel in a spiral arrangement optimized for substrate transfer across the AAA+ domains, thereby establishing a continuous path for polypeptide translocation.
Organizational Affiliation:
Department of Biological Chemistry, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan, USA.