Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Choy, M.S., Li, Y., Machado, L.E., Kunze, M.B., Connors, C.R., Wei, X., Lindorff-Larsen, K., Page, R., Peti, W.(2017) Mol Cell 65: 644-658.e5
- PubMed: 28212750
- DOI: https://doi.org/10.1016/j.molcel.2017.01.014
- Primary Citation of Related Structures:
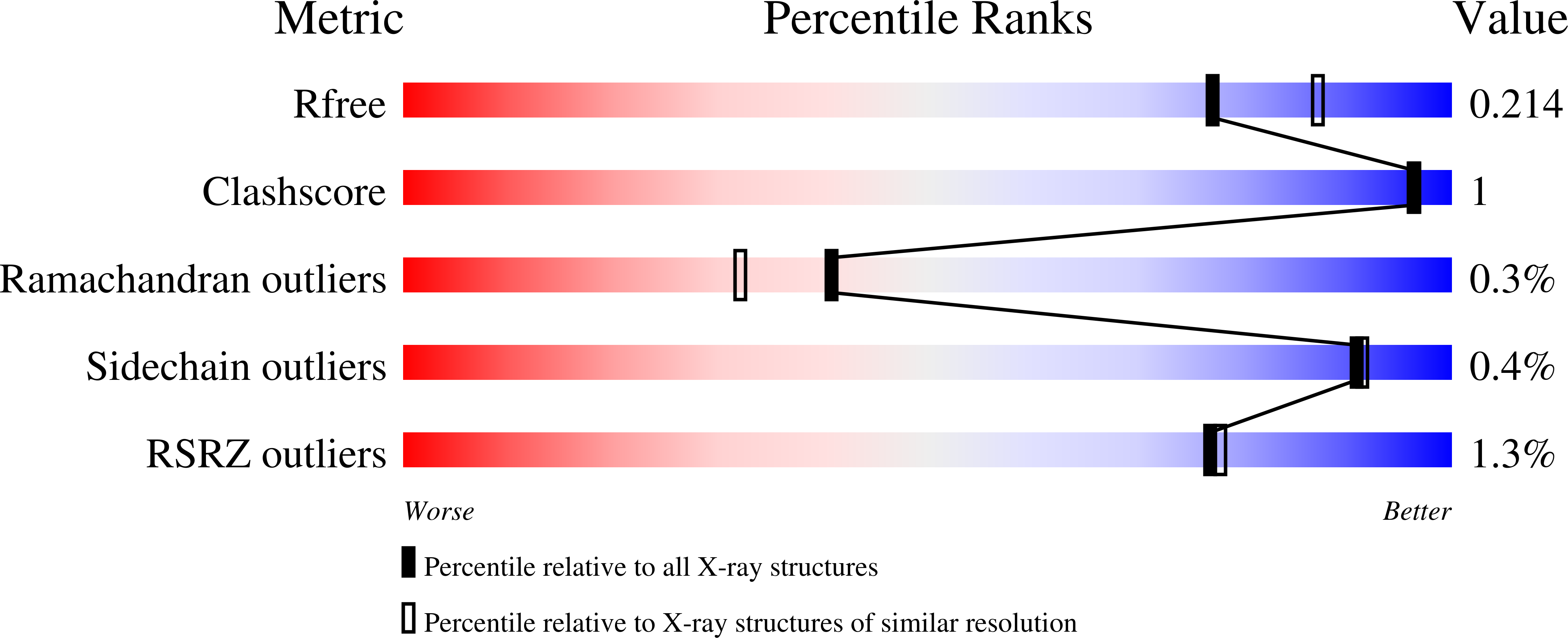
5K9V, 5K9W, 5KA0, 5KA1, 5KA2, 5KA3, 5KA4, 5KA7, 5KA8, 5KA9, 5KAA, 5KAB, 5KAC, 5KAD - PubMed Abstract:
Protein function originates from a cooperation of structural rigidity, dynamics at different timescales, and allostery. However, how these three pillars of protein function are integrated is still only poorly understood. Here we show how these pillars are connected in Protein Tyrosine Phosphatase 1B (PTP1B), a drug target for diabetes and cancer that catalyzes the dephosphorylation of numerous substrates in essential signaling pathways. By combining new experimental and computational data on WT-PTP1B and ≥10 PTP1B variants in multiple states, we discovered a fundamental and evolutionarily conserved CH/π switch that is critical for positioning the catalytically important WPD loop. Furthermore, our data show that PTP1B uses conformational and dynamic allostery to regulate its activity. This shows that both conformational rigidity and dynamics are essential for controlling protein activity. This connection between rigidity and dynamics at different timescales is likely a hallmark of all enzyme function.
Organizational Affiliation:
Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, RI 02912, USA.