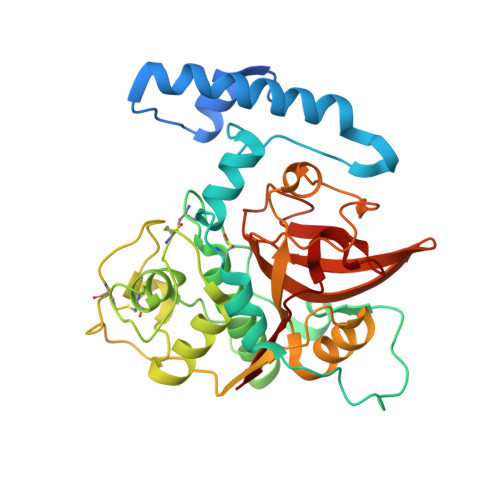
The structure of the mite allergen Blo t 1 explains the limited antibody cross-reactivity to Der p 1.
Meno, K.H., Kastrup, J.S., Kuo, I.C., Chua, K.Y., Gajhede, M.(2017) Allergy 72: 665-670
- PubMed: 27997997
- DOI: https://doi.org/10.1111/all.13111
- Primary Citation of Related Structures:
5JT8 - PubMed Abstract:
The Blomia tropicalis (Blo t) mite species is considered a storage mite in temperate climate zones and an important source of indoor allergens causing allergic asthma and rhinitis in tropical and subtropical regions. Here, we report the crystal structure of one of the allergens from Blo t, recombinant proBlo t 1 (rproBlo t 1), determined at 2.1 Å resolution. Overall, the fold of rproBlo t 1 is characteristic for the pro-form of cysteine proteases from the C1A class. Structural comparison of experimentally mapped Der f 1/Der p1 IgG epitopes to the same surface patch on Blo t 1, as well as of sequence identity of surface-exposed residues, suggests limited cross-reactivity between these allergens and Blo t 1. This is in agreement with ELISA inhibition results showing that, although cross-reactive human IgE epitopes exist, there are unique IgE epitopes for both Blo t 1 and Der p 1.
Organizational Affiliation:
Global Research, ALK-Abelló A/S, Hørsholm, Denmark.